Figure 12.
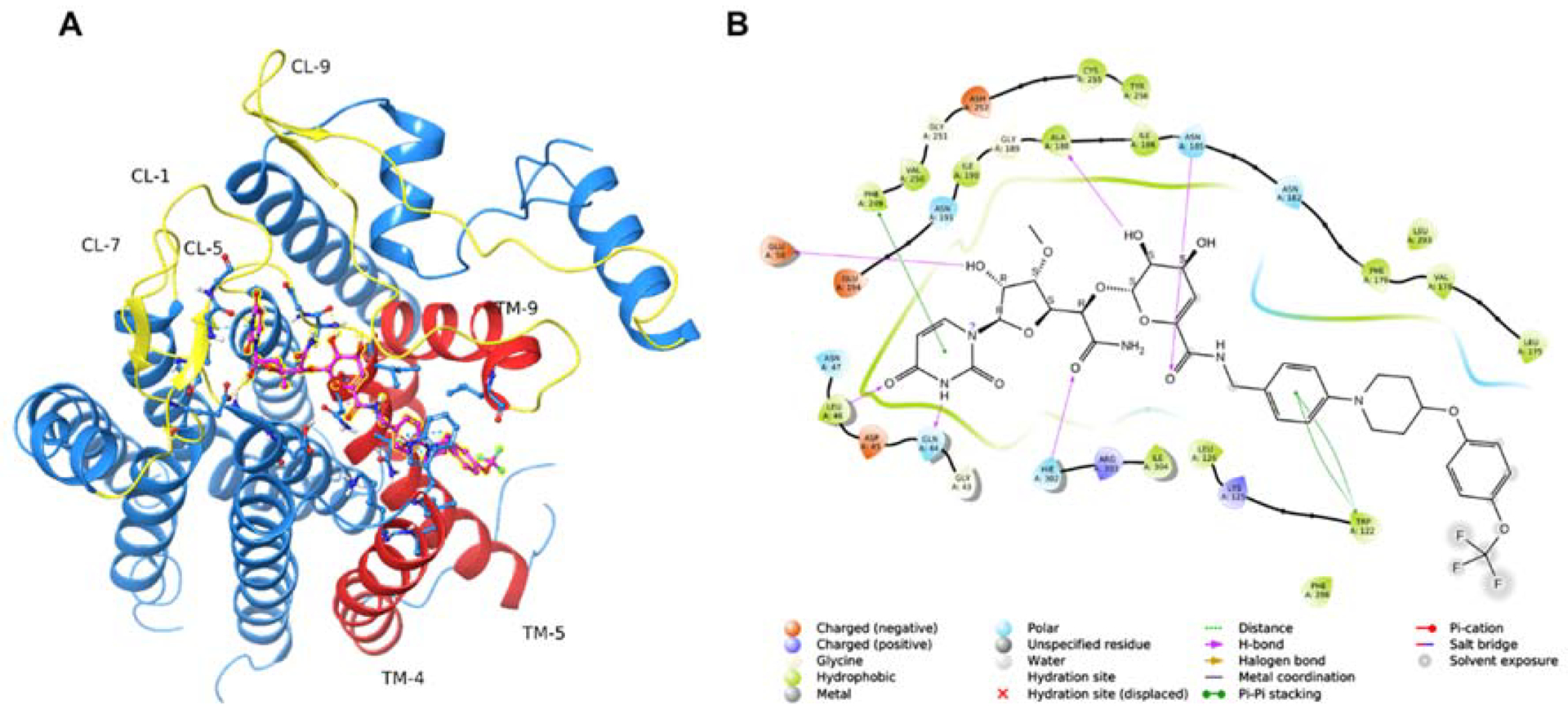
Modeling CPPB-DPAGT1 Interaction to Design New Inhibitor Molecules.a
aDocking studies were performed using the human DPAGT1 with bound tunicamycin (PDB: 6BW6) (PMID 29459785). The biological unit was downloaded and prepared using the Protein Preparation Wizard of the Maestro Small Molecule Drug Discovery Suite (Schrödinger, LLC). The docking receptor grid was prepared using Schrödinger’s Glide program. (PMID 15027866, PMID 15027865). CPPB was built and prepared for docking using the LigPrep program using default settings (Schrödinger, LLC). A: Predicted binding pose of CPPB (highlighted ball & stick model) into DPAGT1. Key active site loops (yellow) and transmembrane regions (orange) are indicated. B: 2D ligand interaction diagram of the docked CPPB-DPAGT1 complex with key predicted interactions shown.
CL: cytoplasmic loop.; TM: transmembrane segment
