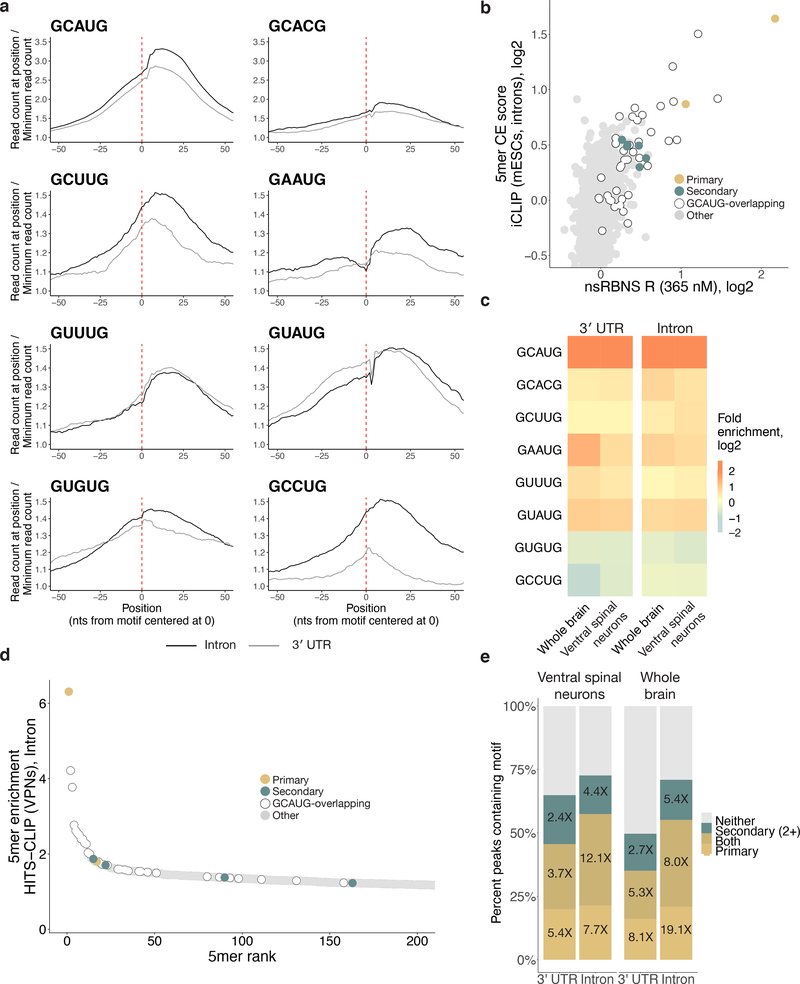
Figure 3. Rbfox proteins bind secondary motifs in vivo.
a. Primary and secondary motif reads peak near 0 in a metaplot centered at the motif in introns (black) and 3' UTRs (grey) in RBFOX2 iCLIP data27 (GEO GSE54794). iCLIP reads containing the motif of interest (see Methods for read counts) were aligned with position one of the pentamer at 0 and normalized to the minimum read count in an 80-nt window (50-nt window shown). Y-axis range was reduced for secondary motifs. b. Correlation of intronic iCLIP- and nsRBNS-enriched 5mers (n = 1024). Secondary motifs indicated in teal, primary motifs indicated in gold. Grey dots indicate “hitchhiking” motifs that overlap the primary motif GCAUG by at least three bases but do not have intrinsic Rbfox affinity. c. RBFOX1 HiTS-CLIP data8,14 (SRA SRP128054, SRP035321) enrichments for primary motifs and secondary six motifs in both 3' UTRs (left, n = 2963 and 989) and introns (right, n = 847 and 1431) relative to transcriptomic frequencies in mouse whole brain and ventral spinal neurons, respectively. Enrichment was calculated based on the 5mer composition of 100-base CLIP peak regions centered around the apex of the CLIP peak relative to 5mer composition of the transcriptomic region. d. Four secondary motifs (GCUUG, GAAUG, GUUUG, GUAUG) are indicated among the top 200 highly enriched 5mers derived from intronic HiTS-CLIP peaks from mouse ventral spinal neurons. Primary motifs in gold, secondary motifs in teal. Peaks calculated as above. e. High-confidence CLIP peaks in two different Rbfox1 HiTS-CLIP datasets (SRA SRP128054, SRP035321) in ventral spinal neurons8 and mouse whole brain cells14 attributable to primary (light gold), or four secondary (teal; GCUUG, GAAUG, GUUUG, and GUAUG), or both (dark gold) motifs in both 3' UTRs (n = 15487 and n = 24972, respectively) and introns (n = 4800 and n = 1519, respectively). Fold enrichments above transcriptomic background are indicated. Peaks containing neither primary nor two or more secondary motifs are shown in grey.