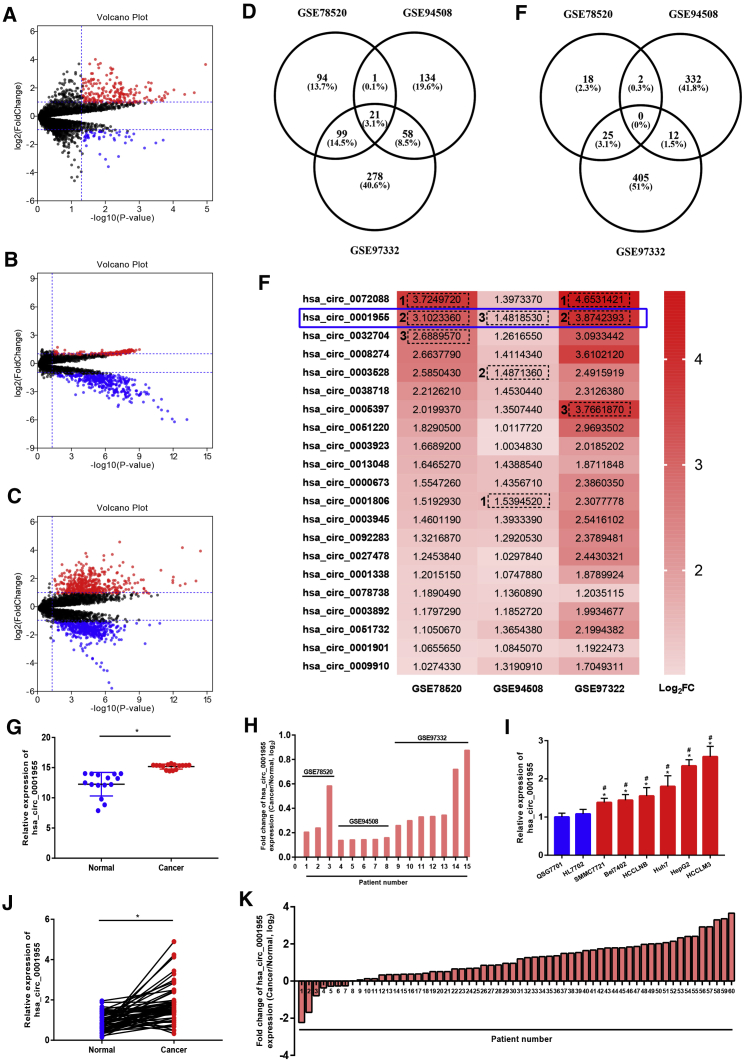
Figure 1.
Identification of hsa_circ_0001955 as the Most Potential circRNA in HCC
(A) The volcano plot of DECs in GSE78520. (B) The volcano plot of DECs in GSE94508. (C) The volcano plot of DECs in GSE97332. The red and blue dots represented upregulated DECs and downregulated DECs with statistical significance (|FC| > 2 and p < 0.05), respectively. (D) The DECs were commonly upregulated in GEO: GSE78520, GSE94508, and GSE97332. (E) The DECs were commonly downregulated in GEO: GSE78520, GSE94508, and GSE97332. (F) The heatmap of 21 DECs that were commonly upregulated in GEO: GSE78520, GSE94508, and GSE97332. (G) hsa_circ_0001955 expression was significantly increased in HCC tissues compared with normal tissues from GEO: GSE78520, GSE94508, and GSE97332. ∗p < 0.05. (H) The expression change (cancer/normal) of hsa_circ_0001955 in 15 HCC patients from GEO: GSE78520, GSE94508, and GSE97332. (I) hsa_circ_0001955 expression in HCC cell lines was higher than that in normal liver cell lines. ∗p < 0.05 (compared with QSG7701); #p < 0.05 (compared with HL7702). (J) hsa_circ_0001955 was markedly upregulated in HCC tissues compared with normal tissues. ∗p < 0.05. (K) The expression change (cancer/normal) of hsa_circ_0001955 in 60 HCC patients.