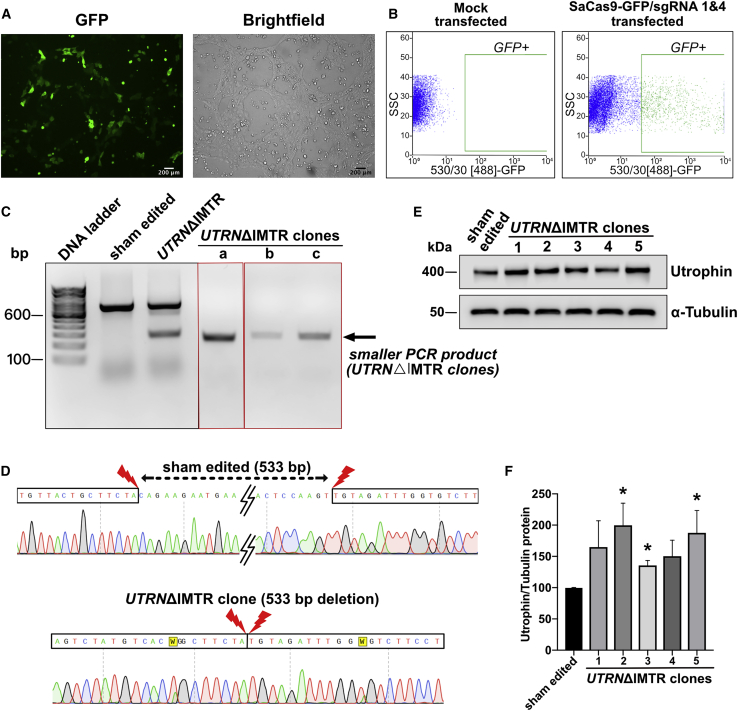
Figure 3.
UTRNΔIMTR Genome Editing in DMD-hiPSCs and Utrophin Protein Upregulation in UTRNΔIMTR Clones
(A) Fluorescence and bright-field microscopy images showing GFP expression in DMD-hiPSCs transfected with SaCas9-GFP/sgRNA1 and 4. Scale bar, 200 μm. (B) FACS sorting of GFP-positive DMD-hiPSCs gated against mock transfected DMD-hiPSCs. (C) Genomic DNA PCR gel from clonally selected, genome edited, DMD-hiPSC lines shows a 267 bp band from UTRNΔIMTR gene (UTRNΔIMTR clones a, b, and c). Whereas genomic DNA PCR from the sham edited (cells transfected with only SaCa9 and no sgRNA) cells shows only unedited 800 bp band. (D) DNA Sequencing of PCR product from UTRNΔIMTR clone shows precise (533 bp) deletion of IMTR compared to sham edited clone. The W in the chromatogram stands for T/A. (E) Representative western blot shows expression of utrophin in DMD-hiPSC sham edited and UTRNΔIMTR clones. α-tubulin was used as loading control. (F) Densitometric analysis of utrophin western blot to quantify utrophin upregulation. Bands were densitometrically quantified and utrophin normalized to α-tubulin. Error bars represent mean ± SEM (n = 4). Difference in utrophin expression between clones were statistically analyzed by the Mann-Whitney test (∗p ≤ 0.05). Significant increase in utrophin expression was observed in UTRNΔIMTR clones 2, 3, and 5 compared to sham edited clones with p value 0.028.