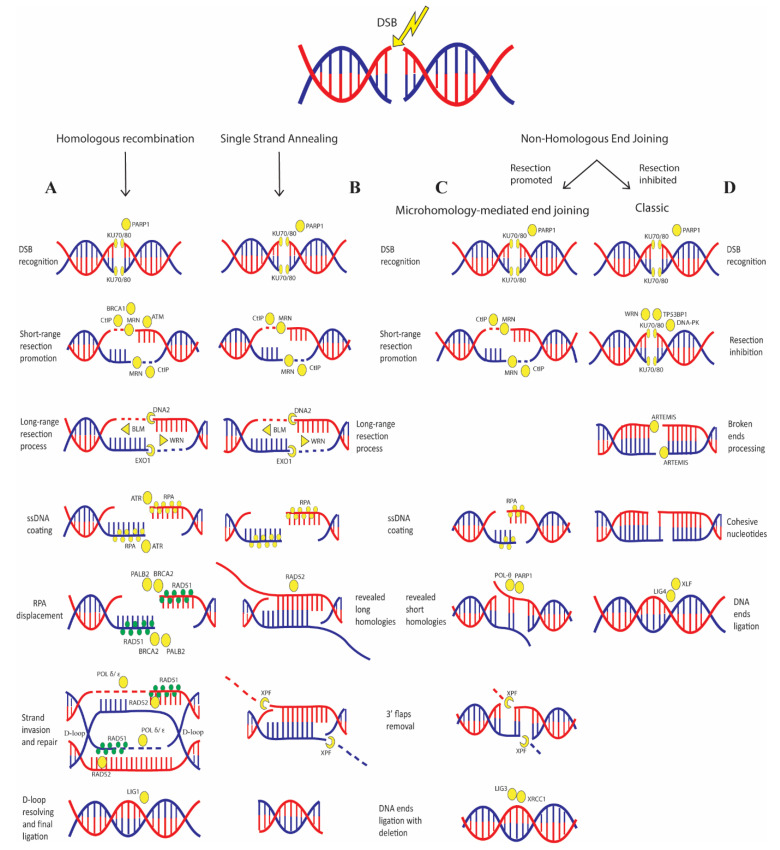
Figure 2.
Several DSB repair pathways can be used depending on cell cycle phase and presence of homology on DNA strand. (A) In the S/G2 phase, when the homologous chromatid is present, HR error-free DSB repair pathway is favored: initial resection performed by the combined action of CtIP, MRN complex and BRCA1, promotes this pathway and allow access of long-range resection nucleases DNA2 and EXO1. WRN and BLM helicases unwind the two DNA strands; the ssDNA formed is rapidly stabilized by RPA and bound by ATR. Thereafter, RPA is displaced by RAD51 aided by BRCA2/PALB2 activity. RAD51 promotes search and invasion of complementary strand and RAD52 determines interaction between the complementary strands; finally, POL δ/ε re-synthetizes the damaged strand and LIG1 performs the final ligation. (B) Similar to HR, SSA requires long range resection of the DSB, but differently from HR this resection process exposes long regions of homology; DNA strands can bind together, also favored by the activity of RAD52, the overhanging 3′ flaps are removed by XLF nucleases and the DNA ends are ligated resulting in long deletion. (C) MMEJ pathway has several steps in common with SSA; the differences are that there is no long-range resection and that homology between strands are short (few base pairs). Short homology is revealed by the activity of Pol θ-associated helicase together with PARP1 and, as in SSA, the 3′flaps are removed by XLF nuclease; in the last step, XRCC1 and LIG3 catalyze DNA ends ligation resulting in short deletion. (D) c- NHEJ is favored by resection inhibition performed by Ku70/80 heterodimer and several other factors modulated by TP53BP1. Ku70/80 activates DNA-PK kinase which recruits ARTEMIS nucleases to the DSB; this promotes the processing of the broken ends until cohesive nucleotides are found, then XLF and LIG4 catalyze the final ligation.