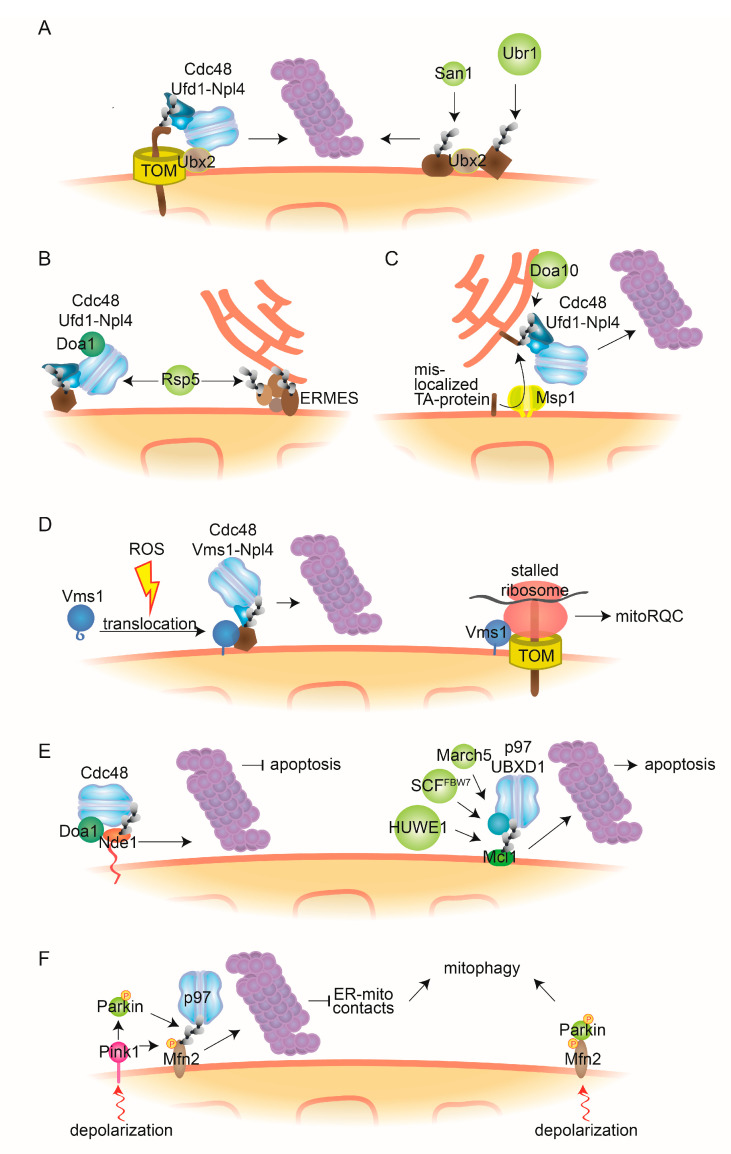
Figure 3.
Cdc48-dependent MAD and mitochondrial quality control mechanisms. (A) Ubx2-dependent MAD. The Cdc48-cofactor Ubx2 partially localizes to the OMM where it facilitates the degradation of ubiquitylated mitochondrial proteins bound to the TOM channel, dependent on the Cdc48-Ufd1-Npl4 complex. Further, Ubx2 recognizes mitochondrial substrates that are ubiquitylated by the quality control E3 ligases Ubr1 and San1. (B) Doa1-dependent MAD. The Cdc48-Ufd1-Npl4 complex, together with the cofactor Doa1/Ufd3, assists proteasomal degradation of OMM proteins ubiquitylated by E3 ligase Rsp5. Additionally, Rsp5 can ubiquitylate the ERMES components Mdm12 and Mdm34. (C) ERAD-assisted degradation. The AAA-ATPase Msp1 facilitates the retro-translocation to the ER of TA-proteins mistargeted to the OMM. After ubiquitylation by the ER-bound E3 ligase Doa10, Cdc48-Ufd1-Npl4 engages the canonical ERAD pathway. (D) Vms1-dependent MAD. Oxidative stress, such as reactive oxygen species (ROS), enables translocation to mitochondria of the Cdc48 cofactor Vms1, where it recruits Cdc48 and Npl4 to assist proteasomal degradation of ubiquitylated mitochondrial substrates. Additionally, Vms1 recognizes stalled ribosomes associated to TOM channels and facilitates release of aberrant nascent peptides, by mitoRQC. (E) Cdc48/p97 in apoptosis. Accumulation of the cytosol-exposed topomer of Nde1 leads to induction of mitophagy. Cdc48 recognizes ubiquitylated forms of this topomer, dependent on Doa1, and facilitates its degradation, thereby preventing apoptosis (left). p97 and its cofactor UBXD1 facilitate proteasomal turnover of the antiapoptotic protein Mcl1, ubiquitylated by the E3 ligases HUWE1, SCFFBW7 or MARCH5, thereby inducing apoptosis (right). (F) p97 in mitophagy. Upon mitochondrial depolarization, PINK1 accumulates at the OMM, where it phosphorylates Parkin, leading to its activation. Parkin-mediated ubiquitylation and p97-mediated degradation of Mfn2 induces mitophagy, e.g., via reduction of ER-mitochondria contacts (left). Alternatively, phosphorylated Mfn2 serves as an adaptor for Parkin, enabling mitophagic signalling (right). MitoRQC, mitochondrial ribosomal quality control; ERMES, ER-mitochondria encounter structure; TOM, translocase of the outer mitochondrial membrane complex.