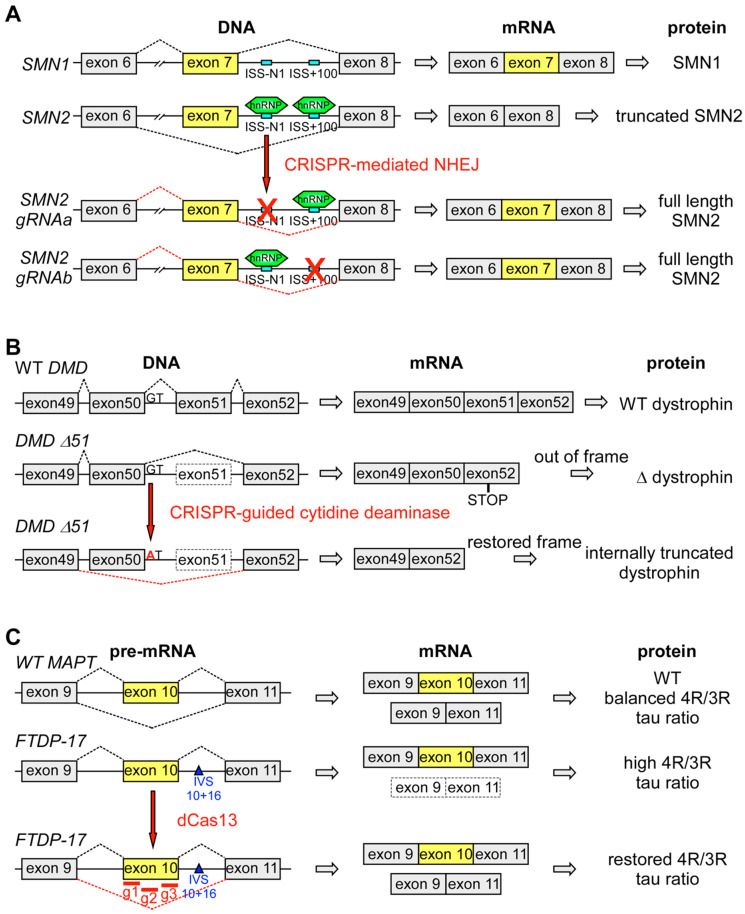
Figure 2.
CRISPR-based splicing therapies (A) CRISPR-mediated non-homologous end joining (NHEJ) disrupts intronic splicing silencer (ISS) sites in intron 7 of SMN2 preventing binding of hnRNPs, thus favoring inclusion of exon 7, normally not present in SMN2. This helps compensate for mutated SMN1. (B) Deletion of exon 51 of DMD leads to a frameshift resulting in a truncated nonfunctional dystrophin. CRISPR-guided cytidine deaminase mutates the 5′ splice site of exon 50, leading to skipping of exon 50, which restores the reading frame. The resulting internally truncated dystrophin is able to restore partially WT function. (C) AS of MAPT exon 10 leads to formation of 4R and 3R tau isoforms. The FTDP-17-associated IVS 10 + 16 mutation results in increased exon 10 inclusion and higher 4R tau levels. Expression of dCas13 together with three gRNAs targeting the exon 10 splice acceptor site and two putative exonic splicing enhancers (ESEs) promotes skipping of exon 10, thus restoring a balanced 4R/3R tau ratio. Schemes adapted from [149,161,164].