Abstract
Background
Osteosarcoma (OS) is a highly aggressive bone malignancy that is mostly diagnosed in children and young adults. Increasing evidence indicates that the transcription factor Forkhead Box M1 (FoxM1) plays a key role in the pathogenesis of various tumors. However, the function of FoxM1 in OS has not been clearly elucidated.
Methods
In the present study, we first analyzed the expressions of FoxM1 in human OS and myositis ossificans (MO, included as a control) tissues by immunohistochemistry. To investigate the functional significance of FoxM1 in OS tumorigenesis, we examined the effects of FoxM1 downregulation in MG-63 and HOS-MNNG cells by either short hairpin RNA (shRNA)-mediated gene silencing or treatment with thiostrepton, a specific FoxM1 inhibitor.
Results
FoxM1 was detected in 82.1% (55/67) of OS vs only 10% (2/20) of MO samples. High expressions of FoxM1 were also detected in three human OS cell lines (HOS-MNNG, MG-63, and U-2OS). FoxM1 downregulation significantly reduced MG-63 and HOS-MNNG cell proliferation, migration, and invasion as well as cell cycle arrest in the G2/M phase and increased apoptotic cell death.
Conclusion
The present study demonstrated the critical role of FoxM1 in the pathogenesis of OS. Therefore, FoxM1 may serve as a potential therapeutic target for the treatment of OS.
Keywords: osteosarcoma, FoxM1, thiostrepton, shRNA, tumorigenesis
Introduction
Osteosarcoma (OS) is the most common type of primary malignant tumor, and it occurs most frequently in children and young adults. The current standard treatment with multi-agent neoadjuvant chemotherapy, followed by surgical removal of the primary tumor can often achieve a long-term survival rate of 65–70%.1 However, little progress has been made in improving patient survival since the mid-1980s,2 highlighting the need for developing novel therapeutic agents to combat this aggressive malignancy.
Forkhead Box M1 (FoxM1) is a transcriptional factor of the forkhead box family that activates the transcription of genes regulating cell cycle progression.3 FoxM1 is ubiquitously expressed in actively proliferating tissues and has a crucial role in embryonic development and adult tissue homeostasis.4–6 In cancer cells, FoxM1 is phosphorylated by the cyclin-dependent kinases 4 and 6 (CDK4/6) to maintain the expression of G1/S-phase genes and protect cells from senescence.3 Recent studies have linked elevated expression of FoxM1 to the initiation, progression, drug resistance, and poor diagnosis of various cancers,7 especially those commonly diagnosed in pediatric patients, such as Ewing sarcoma8 and medulloblastoma.9 In human OS cells, FoxM1 has been reported to be phosphorylated by the cyclin E-CDK2/Raf-MEK-ERK cascade, followed by nuclear translocation to allow transcriptional activation of genes promoting cell cycle progression.9 Nonetheless, the clinical significance and functional role of FoxM1 in human OS remain largely unknown.
In the present study, we investigated the expression of FoxM1 in human OS tissues and cell lines. We further assessed the effects of FoxM1 downregulation, either by short hairpin RNA (shRNA)-mediated gene silencing or by treatment with the FoxM1 inhibitor thiostrepton, on MG-63 and HOS-MNNG OS cell proliferation, apoptosis, migration, and invasion in vitro.
Methods
Cell Culture and Reagents
The human OS cell lines HOS-MNNG, MG-63, U2OS and human embryonic kidney cells 293T were purchased from the American Type Culture Collection (Manassas, VA, USA) and cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco) supplemented with 10% fetal bovine serum (FBS; Gibco). The control osteoblast cell line hFOB1.19 was purchased from iCell Bioscience Inc (Shanghai, China) and cultured in DMEM/F12 with 0.3mg/mL G418 at 34°C in a 5% CO2 humidified incubator. Antibodies against FoxM1 and β-actin were purchased from Abcam (Ab175798) and Abmart (BM0627) (Berkeley Heights, NJ, USA), respectively. Thiostrepton was obtained from Sigma (CAS 1393-48-2). The compound was dissolved in dimethyl sulfoxide (DMSO) and stored at −20°C. The MTT assay and the trypan blue exclusion assay were used to assess cell viability and proliferation.
Tumor Samples and Immunohistochemical Staining
Biopsy samples collected from 67 OS and 20 myositis ossificans (MO) patients prior to treatment at the First Affiliated Hospital of Anhui Medical University (Hefei, China) from January 2012 to December 2016 were included in this study. This study was approved by the Ethics Committee of Anhui Medical University. This study was conducted in accordance with the Declaration of Helsinki. All the patients gave their written informed consent. Of the 67 OS patients, 36 were males and 31 were females. The average age was 24.5 years old (range 7 to 46 years). All cases were high-grade conventional OS (40 osteoblastic, 19 fibroblastic, 7 chondroblastic, and 1 telangiectatic). Tumors were located in the tibia or femur in 41 cases and in the humerus, rib, or other sites in the other 26 cases. All OS samples contained an adequate number of neoplastic cells, according to the histological analysis performed by two bone pathologists (Y.P. Cai and L.Y. Cao). For antigen retrieval, slides were incubated in 0.01 M boiling citrate buffer (pH 6.0) for 10 min. The slides were subsequently incubated with FoxM1 antibody (1:200) at 4°C overnight. Immunohistochemical staining was performed, and immunoreactivity was graded, as described previously.10 In brief, FoxM1 expression was scored as 0 for negative, 2–3 for weakly positive (1+), 4–6 for moderately positive (2+), and 7–8 for strongly positive (3+).
FoxM1 Silencing by shRNA Transfection
PLKO lentiviral vectors containing a FoxM1-targeting shRNA (FoxM1-sh1 or FoxM1-sh2) or a negative control shRNA (NTC) were purchased from Sigma-Aldrich. The following shRNA sequences were used: FoxM1-sh1, CCGGGCCCAACAGGAGTCTAATCAACTCGAGTTGATTAGACTCCTGTTGGGCTTTTT; FoxM1-sh2, CCGGCGCCGGAACATGACCATCAAACTCGAGTTTGATGGTCATGTTCCGGCGTTTTT; NTC, CCGGCAACAAGATGAAGAGCACCAACTCGAGTTGGTGCTCTTCATCTTGTTGTTTTT. The FoxM1-shRNA or control lentivirus were produced by co-transferring the plasmids with VSVG and Δ8.9 to HEK-293T cells. OS cells were seeded on a 6-well plate around 60% confluency before infection. Filtered viral supernatant and polybrene were added to the final concentration of 8μg/mL. Cells were incubated at 37°C overnight and then selected by 2 μg/mL puromycin to generate stable cell lines. The surviving cells were used as stable mass transfectants.
Quantitative Real-Time PCR (qRT–PCR)
Total RNA was isolated using Trizol (Invitrogen) and reversely transcribed using the iScript cDNA Synthesis Kit (Bio-rad) following DNase (Ambion) treatment. qRT-PCR was performed using SYBR Green Master Mix (Vazyme) on a Bio-Rad iCycler. The relative levels of FoxM1 in all samples were normalized to 18S rRNA. The following primer sequences were used: FoxM1, 5ʹ-TGCAGCTAGGGATGTGAATCTTC-3ʹ (forward), 5ʹ-GGAGCCCAGTCCATCAGAACT-3ʹ (reverse); 18S, 5ʹ-CGCTACTACCGATTGGATGG-3ʹ (forward), 5ʹ- AGTTCGACCGTCTTCTCAGC-3ʹ (reverse). The amplification was performed in triplicate with reaction conditions beginning at 95°C for 10 min, then 95°C for 10 sec and 60°C for 30 sec for 40 cycles, followed by 95°C for 15 sec and 60°C for 60 sec. The fold change in expression was calculated based on the crossing point (Cq), where ∆Cq = Cttarget – Ct18S and ∆ (∆Cq) = ∆CqControl – ∆CqIndicated condition.
Western Blotting Analysis
Cells were lysed with radioimmunoprecipitation buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 5 mM EDTA, 0.1% sodium dodecyl sulfate (SDS), and 1% NP-40) supplemented with a protease inhibitor cocktail. Proteins in the supernatants were quantified, separated by SDS-polyacrylamide gel electrophoresis, and immunoblotted with a primary anti-FoxM1 antibody (1:1000) and a horseradish peroxidase-conjugated secondary antibody (Bio-Rad). The signals were detected using Western ECL Substrate (Bio-Rad), according to manufacturer’s instructions. β-actin (1:1000) served as a loading control.
Cell Viability and Proliferation Assays
To examine cell viability, 3000 cells were seeded in 96-well plates and cultured overnight. The cells were subsequently treated with thiostrepton at different concentrations for 48 h. The half-maximal inhibitory concentration (IC50) values of thiostrepton were determined based on the cell viability measured by the MTT assay. To evaluate cell proliferation, 2 × 104 cells were seeded in 12-well plates and cultured in the presence or absence of 4 μM thiostrepton for up to 5 days. Cells stably expressing a FoxM1-targeting shRNA or NTC were cultured for up to 5 days. Cell proliferation was determined by the trypan blue exclusion assay.
Cell Cycle Analysis
Cells were counted (1 × 105 cells in each well) and seeded into six-well culture plates for subsequent analysis of the cell cycle. The medium was substituted after 24 h with DMEM containing a supplement of 5% FBS to make the cells dormant after 24 h. And then indicated treatments were applied to cells for another 24 h. Cells were harvested by trypsinization, washed twice with phosphate-buffered saline containing 5% FBS, and fixed in 70% ethanol and stored in a freezer for 2 h to complete fixation. The cells were subsequently stained with 20 µg/mL propidium iodide (PI) containing 20 µg/mL DNase-free RNase and then incubated for 20 min under light-free conditions at room temperature. Finally, analysis of the cell cycle was carried out, ≥10,000 cells were assessed with a FACS can flow cytometer (Becton Dickinson, Mountain View, CA, USA) and Flow Jover. 7.1.0 (Tree Star, Ashland, OR, USA).
Apoptosis Assay
Cells were treated with 4 μM thiostrepton or infected with lentiviral vectors containing a FoxM1-targeting shRNA or NTC for 48 h. Apoptosis was measured by flow cytometry using the Annexin V-PI Apoptosis Detection Kit (Bestbio), according to manufacturer’s instructions.
In vitro Cell Migration and Invasion
To evaluate cell invasion, 2 × 105 MG-63 cells or 1 × 105 HOS-MNNG cells were placed in serum-free DMEM in the presence or absence of 4 μM thiostrepton in the upper chamber of Matrigel-coated 24-well Transwell plates (Corning Inc., Corning, NY, USA). The lower chamber was filled with 600 μL of DMEM containing 20% FBS. After incubation at 37°C for 24 h, the cells that had migrated to the lower surface of the membrane were fixed in 100% methanol for 30 min, stained with 0.1% crystal violet for 20 min, and counted under a microscope. Migration assays were carried out in uncoated 24-well Transwell plates (Corning), following the otherwise identical experimental procedure. The invasion and migration capacities of MG-63 or HOS-MNNG cells stably expressing a FoxM1-targeting shRNA or NTC were assessed using the same methods in the absence of thiostrepton.
Statistical Analysis
The results are presented as means ± standard deviation. All data were analyzed using SPSS software, version 16.0. Difference between two groups was compared by independent sample Student’s t-test. Comparisons between in more than two groups were used by ANOVA with Bonferroni correction post hoc test. Differences with a two-sided P-value of <0.05 were considered statistically significant.
Results
FoxM1 is Upregulated in Human OS Tissues and Cell Lines
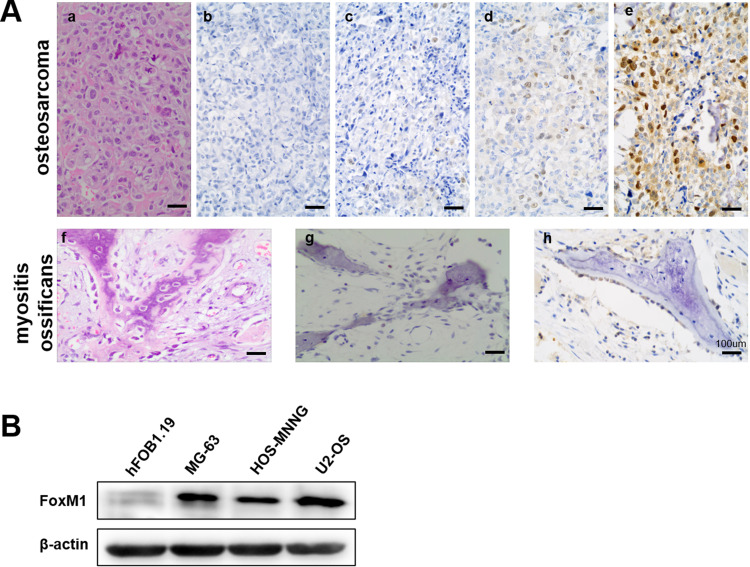
To investigate the role of FoxM1 in OS tumorigenesis, we analyzed the expression of FoxM1 in 67 human OS and 20 human MO tissues by immunohistochemistry (IHC). Our data showed that 82.1% (55/67) of the OS samples exhibited positive FoxM1 staining in the nucleus, with 25 samples showing moderate (2+) and 13 samples showing strong (3+) immunoreactivity (Figure 1D and E and Table 1). In contrast, only 2 out of 20 MO samples showed weak FoxM1 staining in the nucleus (Figure 1H and Table 1). In addition, compared with the human normal cell line hFOB1.19, high FoxM1 protein expression was detected in three human OS cell lines (HOS-MNNG, MG-63, and U-2OS) (Figure 1B). Collectively, our data showed that FoxM1 is highly expressed in OS both in human patient samples and cell lines, indicating that FoxM1 might be a bio-mark of OS.
Figure 1 .
FoxM1 is upregulated in human OS tissues and cell lines. (A) (a, f) Hematoxylin and eosin staining of human OS (a) and MO (f) tissues. (b-e) Immunohistochemical staining for FoxM1 in human OS (b: negative; c: 1+; d: 2+; e: 3+) and MO (g: negative; f: 1+) tissues, scale bars=100um. (B) FoxM1 protein levels in three OS cell lines (HOS-MNNG, MG-63, and U2OS) is higher than in osteoblast cell line hFOB1.19 by western blot analysis. β-actin served as a loading control.
Table 1.
Immunohistochemical FoxM1 Expression in Myositis Ossificans and OS Samples
| Group | Number | FoxM1 Score | Positive Rate | |||
|---|---|---|---|---|---|---|
| 0 | 1+ | 2+ | 3+ | |||
| Myositis ossificans | 20 | 18 | 2 | 0 | 0 | 10.0% |
| OS | 67 | 12 | 17 | 25 | 13 | 82.1% |
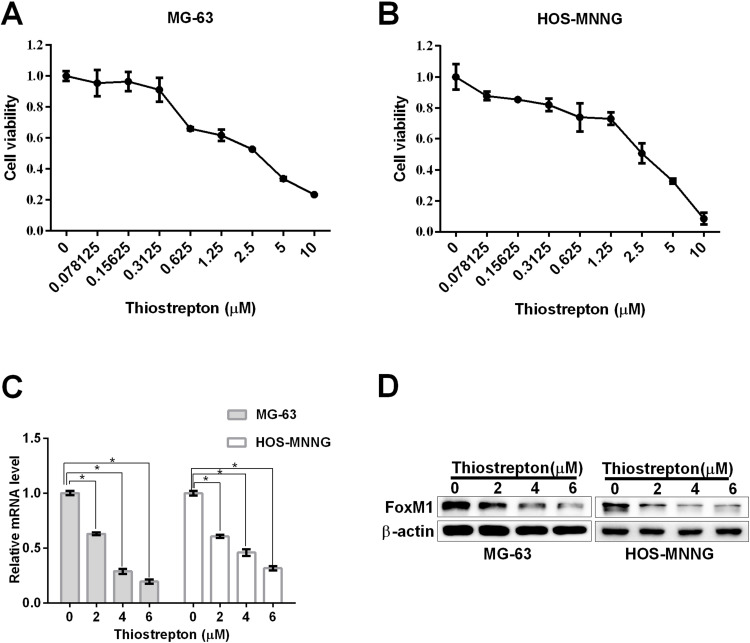
The FoxM1 Inhibitor Thistrepton Reduces OS Cell Viability and the Expression of FoxM1
We next investigated whether targeting FoxM1 affects OS cell viability. We used a FoxM1-specific inhibitor, thiostrepton, and found a dose-dependently reduced cell viability in MG-63 and HOS-MNNG cells, as assessed by the MTT assay after treatment for 48 h (Figure 2A and B). The IC50 values of thiostrepton in MG-63 and HOS-MNNG cells were determined to be 3.3 μM and 4.2 μM, respectively (Figure 2A and B). These data suggested that blocking FoxM1 with its specific inhibitor impairs the cell viability of OS cell lines.
Figure 2 .
The FoxM1 inhibitor thiostrepton reduces OS cell viability and FoxM1 expression. (A, B) MTT assays were performed to determine the IC50 values of thiostrepton in MG-63 (A) and HOS-MNNG (B) cells. The IC50 value of thiostrepton in MG-63 and HOS-MNNG cells was approximately 4 µM. (C, D) FoxM1 protein and mRNA levels in OS cells treated with 2, 4, and 6 M thiostrepton for 48 h by qRT-PCR (C) and western blot analysis (D), respectively. n = 3, *P < 0.05 vs. DMSO.
We next investigated how inhibitor thiostrepton impairs the cell viability of OS cell lines. We detected the mRNA level and protein level of FoxM1 with qRT-PCR and Western blot assay, respectively. Our data showed that the reduced cell viability was accompanied by decreased FoxM1 protein and mRNA expression, as revealed by qRT-PCR (Figure 2C) and Western blot analysis (Figure 2D), respectively, suggesting that inhibition of FoxM1 reduces OS cell viability.
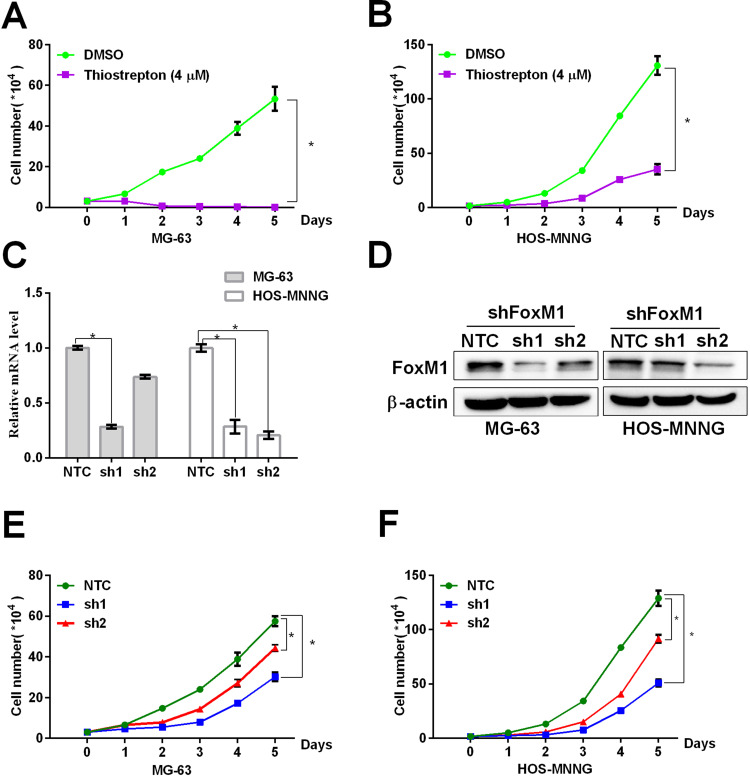
FoxM1 Downregulation by Thiostrepton or shRNA-Mediated Gene Silencing Inhibits OS Cell Proliferation
To investigate the effects of FoxM1 inhibition on OS cell proliferation, we treated the OS cell lines MG-63 and HOS-MNNG with 4 μM thiostrepton or DMSO for up to 5 days, and measured the cell proliferation by the trypan blue exclusion assay. The results showed that thiostrepton significantly inhibited the cell proliferation of both cell lines (Figure 3A and B). To confirm the result that blocking FoxM1 with its inhibitor thiostrepton impairs OS cell proliferation, we transduced OS cell lines MG-63 and HOS-MNNG with FoxM1-specific shRNAs and our data showed that compared with the non-targeting control (NTC), OS cells expressing FoxM1-sh1 or FoxM1-sh2 showed significant decrease of FoxM1 mRNA (Figure 3C) and protein (Figure 3D) expression. And compared with NTC, MG-63 and HOS-MNNG cells expressing FoxM1-sh1 or FoxM1-sh2 exhibited a decreased cell growth rate (Figure 3E and F). These data indicated that FoxM1 downregulation inhibits OS cell proliferation.
Figure 3 .
FoxM1 downregulation by thiostrepton or shRNA-mediated gene silencing inhibits OS cell proliferation. (A, B) MG-63 (A) or HOS-MNNG (B) cells were treated with 4 M thiostrepton or DMSO for up to 5 days. Growth curves were determined by the trypan blue exclusion assay. (C, D) FoxM1 protein and mRNA levels in MG-63 and HOS-MNNG cells expressing FoxM1-sh1 or FoxM1-sh2, compared with NTC, by qRT-PCR (C) and western blot analysis (D), respectively. n = 3, *P < 0.05 vs. DMSO or NTC. (E, F) MG-63 (E) and HOS-MNNG (F) cells expressing FoxM1-sh1, FoxM1-sh2, or NTC were cultured for up to 5 days. Growth curves were determined by the trypan blue exclusion assay. n = 3, *P < 0.05 vs. DMSO or NTC.
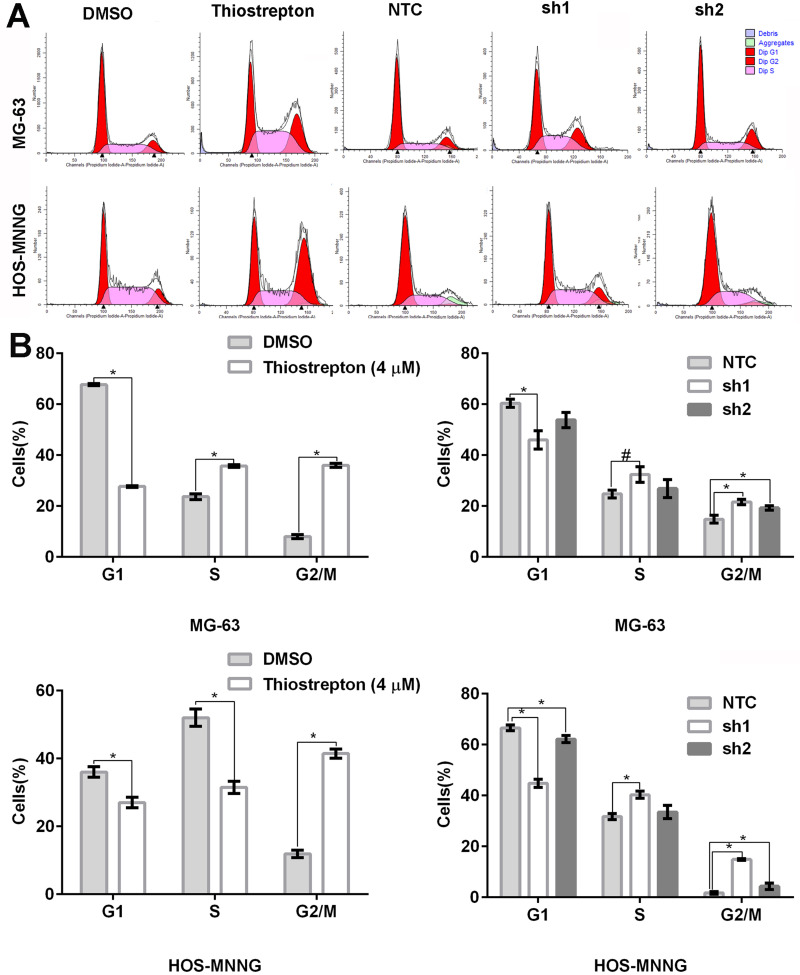
FoxM1 Downregulation by Thiostrepton or shRNA-Mediated Gene Silencing Induces Cell Cycle Arrest in the G2/M Phase
To explore the mechanisms by which FoxM1 downregulation inhibits OS cell proliferation, we assessed the cell cycle distribution by flow cytometry. Compared with the control, MG-63 and HOS-MNNG cells treated with 4 μM thiostrepton exhibited a greater percentage of cells in the G2/M phase (MG-63: 36.0% (thiostrepton) vs 8.0% (DMSO), HOS-MNNG: 41.5% (thiostrepton) vs 11.9% (DMSO)) (Figure 4A and B). In addition, MG-63 and HOS-MNNG cells expressing FoxM1-sh1 or FoxM1-sh2 displayed a higher G2/M-phase cell population than their respective control (MG-63: 21.6% (FoxM1-sh1) or 19.3% (FoxM1-sh2) vs 14.8% (NTC), HOS-MNNG: 14.9% (FoxM1-sh1) or 4.4% (FoxM1-sh2) vs 1.7% (NTC)) (Figure 4A and B). These data indicated that FoxM1 downregulation by inhibitor thiostrepton or shRNA induces G2/M-phase arrest in OS cells.
Figure 4 .
FoxM1 downregulation induces cell cycle arrest in the G2/M phase. MG-63 and HOS-MNNG cells were treated with 4 µM thiostrepton or DMSO for 48 h or stably transduced with lentiviral vectors carrying FoxM1-sh1, FoxM1-sh2, or NTC. The cell cycle distribution was assessed by flow cytometry. Histograms (A) and quantification of the cell cycle distribution (B) are shown. n = 3, *P < 0.05 vs. DMSO or NTC. #P is 0.05, it is marginally significant.
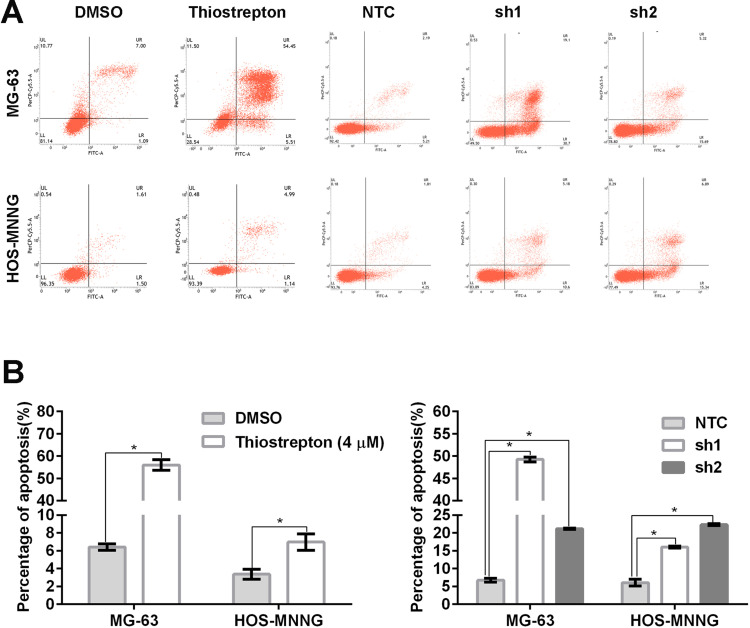
FoxM1 Downregulation by Thiostrepton or shRNA-Mediated Gene Silencing Promotes OS Cell Apoptosis
Next, we evaluated cell apoptosis by flow cytometry using fluorescein-conjugated annexin V and PI double staining. Compared with the control, MG-63 and HOS-MNNG cells treated with 4 μM thiostrepton for 48 h displayed a greater percentage of apoptotic cells (MG-63: 56.0% (thiostrepton) vs 6.4% (DMSO), HOS-MNNG: 7.0% (thiostrepton) vs 3.4% (DMSO)) (Figure 5A and B). In addition, MG-63 and HOS-MNNG cells expressing FoxM1-sh1 or FoxM1-sh2 showed increased cell apoptosis compared with their respective control (MG-63: 49.2% (FoxM1-sh1) or 21.1% (FoxM1-sh2) vs 6.8% (NTC), HOS-MNNG: 16.0% (FoxM1-sh1) or 22.3% (FoxM1-sh2) vs 6.1% (NTC)) (Figure 5A and B). These data indicated that FoxM1 downregulation promotes OS cell apoptosis.
Figure 5.
FoxM1 downregulation promotes OS cell apoptosis. MG-63 and HOS-MNNG cells were treated with 4 µM thiostrepton or DMSO for 48 h or stably transduced with lentiviral vectors carrying FoxM1-sh1, FoxM1-sh2, or NTC. Apoptosis was assessed by flow cytometry using annexin V/PI double staining. Histograms (A) and quantification of apoptosis (B) are shown. n = 3, *P < 0.05 vs. DMSO or NTC.
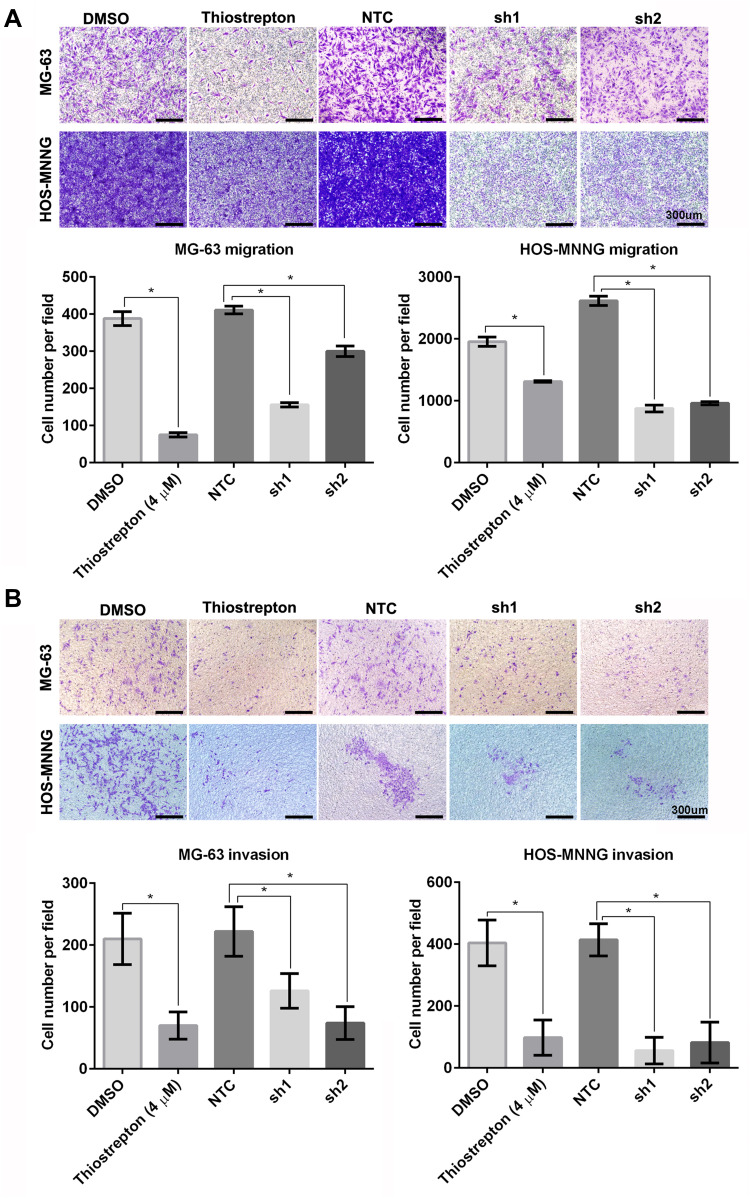
FoxM1 Downregulation by Thiostrepton or shRNA-Mediated Gene Silencing Inhibits OS Cell Migration and Invasion
To investigate the functional significance of FoxM1 in OS tumorigenesis, we examined the effects of FoxM1 downregulation in MG-63 and HOS-MNNG cells. We examined the migration and invasion capacities of OS cells using transwell assays. We found that FoxM1 downregulation by either thiostrepton treatment or shRNA-mediated gene silencing led to dramatic reduced cell migration (Figure 6A) as well as cell invasion (Figure 6B) in both MG-63 and HOS-MNNG cells with significant decreased crystal violet staining. These data demonstrated that FoxM1 downregulation decreases the migration and invasion capacities of OS cells in vitro and FoxM1 may serve as a potential therapeutic target for the treatment of OS.
Figure 6 .
FoxM1 downregulation inhibits OS cell migration and invasion. MG-63 and HOS-MNNG cells were treated with 4 μM thiostrepton or DMSO, or stably transduced with lentiviral vectors carrying FoxM1-sh1, FoxM1-sh2, or NTC. The migration (A) and invasion (B) capacities of these cells were evaluated by Transwell migration and invasion assays, respectively. n = 3, *P < 0.05 vs. DMSO or NTC. Scale bars =300um.
Discussion
The prognosis of patients with recurrent or refractory OS is extremely poor,2,11 highlighting the urgent need for more effective treatments of this grievous malignancy. In the present study, we found that FoxM1, a biomarker for a poor prognosis in many malignancies,12–15 is upregulated in human OS tissues and MG-63 or HOS-MNNG cell lines. These data are consistent with previous reports showing that FoxM1 is overexpressed in a large set of human OS tissues.16 Moreover, we reported FoxM1 is highly expressed in OS in comparison to myositis ossificans. One limitation of our study is that the sample size of 67 OS cases is relatively small; however, considering that osteosarcoma is a rare tumor, it was very difficult to obtain a greater number of clinical OS samples from our hospital. Also, to ensure data quality, we only included samples showing adequate numbers of viable neoplastic cells but no bone tissues, further limiting the number of samples that we could use. We also found that inhibition of FoxM1 by treatment with the inhibitor thiostrepton or shRNA-mediated gene silencing led to decreased OS cell viability and proliferation, suggesting that FoxM1 is involved in OS tumorigenesis.
FoxM1 is a transcription factor that promotes cell proliferation through regulation of genes involved in the G1/S and G2/M phases of the cell cycle.3,17 For example, FoxM1 depletion in U2OS cells dysregulates genes controlling mitosis, such as PLK1, cyclin B1, Cdc25B, Aurora B kinase, and survivin, and consequently causes G2/M-phase arrest and mitotic catastrophe.18 In addition, FoxM1 promotes B-ALL cell proliferation and drug resistance through transcriptional activation of cyclin B1 and Aurora B, two critical regulators of the G2/M phase.19 In alignment with these previous findings, our mechanistic studies showed that FoxM1 downregulation by thiostrepton or shRNA-mediated gene silencing caused G2/M-phase arrest in human MG-63 and HOS-MNNG OS cells. Previous studies have reported that FoxM1 inhibition by thiostrepton induces cell death through caspase-dependent intrinsic and extrinsic apoptotic pathways in breast cancer cells20 and that FoxM1 knockdown in hypopharyngeal squamous cell carcinoma promotes apoptotic cell death.21 In this study, we found that FoxM1 inhibition by thiostrepton or shRNA-mediated gene silencing increased apoptotic cell death in OS cells. These results suggested that FoxM1 promotes OS cell growth by driving entry into mitosis and suppressing apoptosis.
FoxM1 also has been recognized as a master regulator of cancer metastasis through regulation of genes involved in metastatic processes including cell migration and invasion.7,22,23 For instance, FoxM1 depletion in glioma cells reduces cell migration and invasion through downregulation of matrix metalloproteinase-2.24 In addition, FoxM1 transactivates Cav-1 in pancreatic cancer cells to promote epithelial–mesenchymal transition as well as cancer invasion and metastasis.25 In esophageal cancer, FoxM1 promotes tumor metastasis by transcriptionally activating interferon regulatory factor 1 expression.26 In the present study, we found that FoxM1 downregulation by thiostrepton or shRNA-mediated gene silencing attenuated the migration and invasion capacities of MG-63 and HOS-MNNG OS cells in vitro, suggesting a functional role of FoxM1 in OS metastasis. Consistent with our study, Fan et al report found that high expression of FoxM1 is correlated with a poor prognosis.16 However, the absence of mechanistic study is a limitation of our study and further investigations are needed to address this question.
In summary, we demonstrated that FoxM1 contributes to the pathogenesis of OS by driving MG-63 and HOS-MNNG cell proliferation, suppressing cell apoptosis, as well as promoting migration and invasion. Our data suggest that FoxM1 may represent a novel target for the development of effective therapeutics to combat OS. Thiostrepton may represent a novel lead compound for targeted therapy of OS, further in vivo and in vitro investigations are needed to evaluate the efficacy of the FoxM1 inhibitor thiostrepton in OS treatment, either alone or in combination with other agents.
Funding Statement
This work was supported by grants from the National Science Foundation of China (81102041) and the Anhui Provincial Natural Science Foundation (1040606Q17).
Data Sharing Statement
The data associated with the current study are available from the corresponding author YP Cai upon request.
Ethics Approval
This study was approved by the Ethics Committee of Anhui Medical University (approval number: 2019H015) and consents were obtained from the participants.
Disclosure
The authors declare no conflicts of interest.
References
- 1.Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004: data from the surveillance, epidemiology, and end results program. Cancer. 2009;115(7):1531–1543. doi: 10.1002/cncr.24121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Isakoff MS, Bielack SS, Meltzer P, Gorlick R. Osteosarcoma: current treatment and a collaborative pathway to success. J Clin Oncol. 2015;33(27):3029–3035. doi: 10.1200/JCO.2014.59.4895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anders L, Ke N, Hydbring P, et al. A systematic screen for CDK4/6 substrates links FOXM1 phosphorylation to senescence suppression in cancer cells. Cancer Cell. 2011;20(5):620–634. doi: 10.1016/j.ccr.2011.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yao S, Fan LY, Lam EW. The FOXO3-FOXM1 axis: a key cancer drug target and a modulator of cancer drug resistance. Semin Cancer Biol. 2018;50:77–89. doi: 10.1016/j.semcancer.2017.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kelleher FC, O’Sullivan H. FOXM1 in sarcoma: role in cell cycle, pluripotency genes and stem cell pathways. Oncotarget. 2016;7(27):42792–42804. doi: 10.18632/oncotarget.8669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lam EW, Brosens JJ, Gomes AR, Koo CY. Forkhead box proteins: tuning forks for transcriptional harmony. Nat Rev Cancer. 2013;13(7):482–495. doi: 10.1038/nrc3539 [DOI] [PubMed] [Google Scholar]
- 7.Liao GB, Li XZ, Zeng S, et al. Regulation of the master regulator FOXM1 in cancer. Cell Commun Signal. 2018;16(1):57. doi: 10.1186/s12964-018-0266-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christensen L, Joo J, Lee S, Wai D, Triche TJ, May WA. FOXM1 is an oncogenic mediator in Ewing sarcoma. PLoS One. 2013;8(1):e54556. doi: 10.1371/journal.pone.0054556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Priller M, Poschl J, Abrao L, et al. Expression of FoxM1 is required for the proliferation of medulloblastoma cells and indicates worse survival of patients. Clin Cancer Res. 2011;17(21):6791–6801. doi: 10.1158/1078-0432.CCR-11-1214 [DOI] [PubMed] [Google Scholar]
- 10.Wan Y, Zhao WD, Jiang Y, Liu DB, Meng G, Cai YP. Beta-catenin is a valuable marker for differential diagnosis of osteoblastoma and osteosarcoma. Hum Pathol. 2014;45(7):1459–1465. doi: 10.1016/j.humpath.2014.02.022 [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, Yang J, Zhao N, et al. Progress in the chemotherapeutic treatment of osteosarcoma. Oncol Lett. 2018;16(5):6228–6237. doi: 10.3892/ol.2018.9434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nandi D, Cheema PS, Jaiswal N, Nag A. FoxM1: repurposing an oncogene as a biomarker. Semin Cancer Biol. 2018;52:74–84. doi: 10.1016/j.semcancer.2017.08.009 [DOI] [PubMed] [Google Scholar]
- 13.Kuda M, Kohashi K, Yamada Y, et al. FOXM1 expression in rhabdomyosarcoma: a novel prognostic factor and therapeutic target. Tumour Biol. 2016;37(4):5213–5223. doi: 10.1007/s13277-015-4351-9 [DOI] [PubMed] [Google Scholar]
- 14.Intuyod K, Saavedra-Garcia P, Zona S, et al. FOXM1 modulates 5-fluorouracil sensitivity in cholangiocarcinoma through thymidylate synthase (TYMS): implications of FOXM1-TYMS axis uncoupling in 5-FU resistance. Cell Death Dis. 2018;9(12):1185. doi: 10.1038/S41419-018-1235-0 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Zona S, Bella L, Burton MJ, Nestal de Moraes G, Lam EW. FOXM1: an emerging master regulator of DNA damage response and genotoxic agent resistance. Biochim Biophys Acta. 2014;1839(11):1316–1322. doi: 10.1016/j.bbagrm.2014.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fan CL, Jiang J, Liu HC, Yang D. Forkhead box protein M1 predicts outcome in human osteosarcoma. Int J Clin Exp Med. 2015;8(9):15563–15568. [PMC free article] [PubMed] [Google Scholar]
- 17.Wang IC, Chen YJ, Hughes D, et al. Forkhead box m1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol Cell Biol. 2005;25(24):10875–10894. doi: 10.1128/Mcb.25.24.10875-10894.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Major ML, Lepe R, Costa RH. Forkhead box M1B transcriptional activity requires binding of Cdk-cyclin complexes for phosphorylation-dependent recruitment of p300/CBP coactivators. Mol Cell Biol. 2004;24(7):2649–2661. doi: 10.1128/mcb.24.7.2649-2661.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Consolaro F, Basso G, Ghaem-Magami S, Lam EWF, Viola G. FOXM1 is overexpressed in B-acute lymphoblastic leukemia (B-ALL) and its inhibition sensitizes B-ALL cells to chemotherapeutic drugs. Int J Oncol. 2015;47(4):1230–1240. doi: 10.3892/ijo.2015.3139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwok JMM, Myatt SS, Marson CM, Coombes RC, Constantinidou D, Lam EWF. Thiostrepton selectively targets breast cancer cells through inhibition of forkhead box M1 expression. Mol Cancer Ther. 2008;7(7):2022–2032. doi: 10.1158/1535-7163.MCT-08-0188 [DOI] [PubMed] [Google Scholar]
- 21.Chen Y, Liu YF, Ni HS, Ding CJ, Zhang XB, Zhang ZX. FoxM1 overexpression promotes cell proliferation and migration and inhibits apoptosis in hypopharyngeal squamous cell carcinoma resulting in poor clinical prognosis. Int J Oncol. 2017;51(4):1045–1054. doi: 10.3892/ijo.2017.4094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferrer CM, Lu TY, Bacigalupa ZA, Katsetos CD, Sinclair DA, Reginato MJ. O-GlcNAcylation regulates breast cancer metastasis via SIRT1 modulation of FOXM1 pathway. Oncogene. 2017;36(4):559–569. doi: 10.1038/onc.2016.228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang W, Guo ZJ, Yu H, Fan L. MiR-216b inhibits osteosarcoma cell proliferation, migration, and invasion by targeting Forkhead Box M1. J Cell Biochem. 2019;120(4):5435–5443. doi: 10.1002/jcb.27822 [DOI] [PubMed] [Google Scholar]
- 24.Dai B, Kang SH, Gong W, et al. Aberrant FoxM1B expression increases matrix metalloproteinase-2 transcription and enhances the invasion of glioma cells. Oncogene. 2007;26(42):6212–6219. doi: 10.1038/sj.onc.1210443 [DOI] [PubMed] [Google Scholar]
- 25.Huang C, Qiu ZJ, Wang LW, et al. A novel FoxM1-caveolin signaling pathway promotes pancreatic cancer invasion and metastasis. Cancer Res. 2012;72(3):655–665. doi: 10.1158/0008-5472.CAN-11-3102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou YZ, Wang Q, Chu L, et al. FOXM1c promotes oesophageal cancer metastasis by transcriptionally regulating IRF1 expression. Cell Prolif. 2019;52(2):e12553. doi: 10.1111/cpr.12553 [DOI] [PMC free article] [PubMed] [Google Scholar]