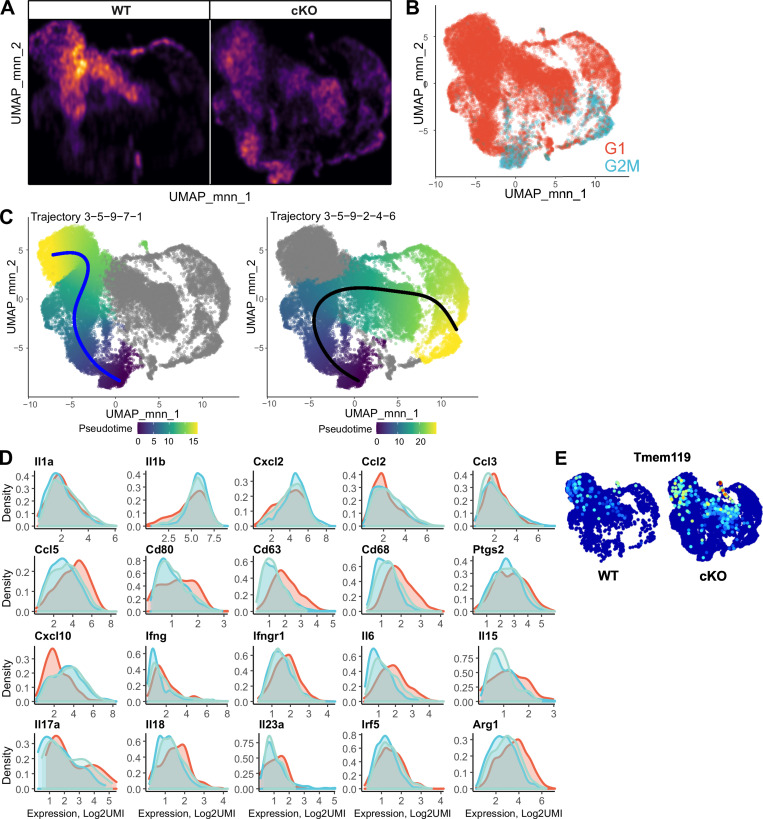
Figure S4.
scRNAseq of spinal cord myeloid cells from WT and GRIP1-cKO mice at EAE DPI20. (A) Bootstrapping analysis of cell densities in the UMAP1–UMAP2 coordinates, stratified by genotypes. 2,000 cells were randomly sampled for each genotype, and the two-dimensional density matrix was calculated for an 800 × 800 binned matrix. The sampling was performed 500 times, and the average density for each bin was computed and plotted. (B) Automated cell cycle stage assignment for 20,376 spinal cord–derived myeloid cells. (C) singleR trajectory inference for 20,376 spinal cord–derived myeloid cells mapped onto a UMAP plot and colored by pseudo-time. Blue and black lines represent fitted PC curves. (D) Expression of MΦ markers in MΦ-like clusters 1 (red), 2 (blue), and 7 (light green). The color of expression profiles corresponds to Louvain clusters in Fig. 6 C. (E) Expression of homeostatic MG markers mapped onto a UMAP plot, stratified by genotypes.