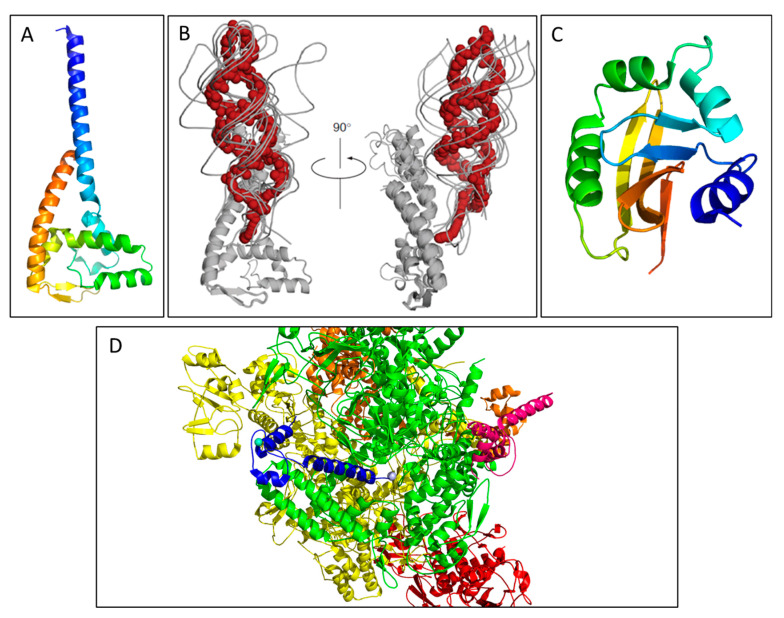
Figure 2.
Structures of proteins with transcription regulatory roles in the F-plasmid tra operon. (A) The crystal structure of a truncated FinO lacking the disordered N-terminal domain (PDB ID: 1DVO). This structure was used for modeling of small angle X-ray scattering (SAXS) data shown in (B), which displays the dynamic conformational ensemble involved in the binding of FinP SLII RNA (red spheres display the average structure of the best cluster of nine complexes, also shown as grey ribbons) to a truncated FinO monomer shown as a grey cartoon [28]. (C) The structurally conserved Per-ARNT-Sim (PAS) domain of TraJ; it is the only region of a TraJ structure currently solved to high resolution (PDB ID: 4KQD). (D) The structure of TraR bound to RNA polymerase (RNAP) in the conformation exhibiting the highest occupancy as determined by cryo-EM. TraR, shown in blue, is seen with its N-terminal helix extending from the secondary channel of the active site in the β-subunit colored green. These residues are shown to interact strongly with the RNAP motif that chelates the Mg2+ ion seen in grey (PDB ID: 6N57). Structural images in Figure 2A–D, Figures 4 and 5 were generated with PyMOL [35]. Figure 2B was adapted with permission from Arthur et al. Mapping interactions between the RNA chaperone FinO and its RNA targets. Nucl. Acid Res. 2011, 39, 4450-63 [28].