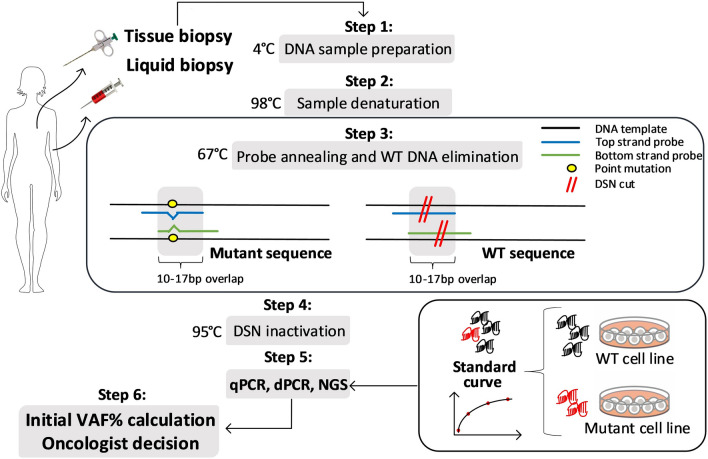
Figure 1.
Workflow of nuclease-based mutant allele enrichment assay and initial VAF prediction. The enrichment protocol is based on duplex specific nuclease (DSN) preferentially degrading perfectly annealed double stranded DNA (dsDNA). DNA sample preparation introducing oligonucleotide probes which overlap the mutation location and are specific to the wild type (WT) sequence (step 1), is followed by sample denaturation (step 2), probe annealing to target DNA sequences and DSN digestion at perfectly annealed WT sequences (step 3) and nuclease inactivation at the end (step 4), thus enriching for mutant DNA sequences. After sample analysis with the user’s preferred method (step 5) initial VAF is calculated from a standard curve of known VAF DNA samples (step 6) and can be used to follow-up patient progression in the clinical setting.