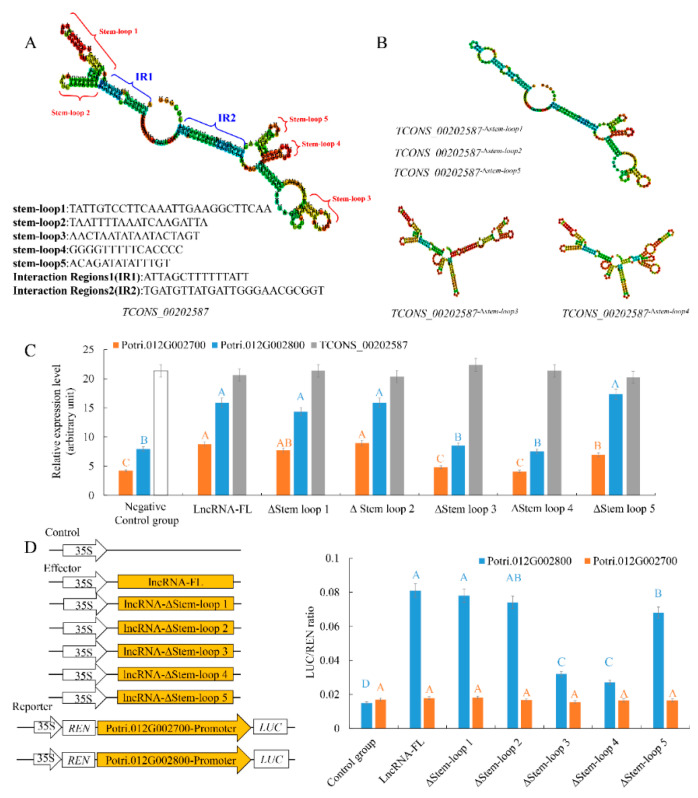
Figure 4.
The secondary structure of TCONS_00202587 regulates the transcript levels of its targets. (A) Schematic diagram of the secondary structure and sequences of TCONS_00202587: Stem-loops 1–5 represent the typical secondary structure of TCONS_00202587. IR 1 and IR2 represent two potential interaction regions binding with the promoter of targets. (B) Schematic diagram of secondary structure mutants: TCONS_00202587-∆stem-loop3 and TCONS_00202587-∆stem-loop4 show significant changes in their secondary structures. (C) Target expression patterns under LDH-NP binding with TCONS_00202587 and its secondary structure mutant: lncRNA-FL represents full-length lncRNA. ∆stem-loop 1–5 represents deletion of stem-loops 1–5, respectively. Negative control group represents LDH-NPs binding with a random lncRNA sequence. (D) Transcriptional activation of TCONS_00202587 and its secondary structure mutant on two targets: The data are mean ±standard deviation (SD) of separate transfections (n = 3). Significantly different genes among the control group, LncRNA-FL, ∆stem-loop 1, ∆stem-loop 2, ∆stem-loop 3, ∆stem-loop 4, and ∆stem-loop 5 were determined using the LSD test. Different colored letters on error bars indicate significant differences in the genes corresponding to that color among groups at p < 0.01.