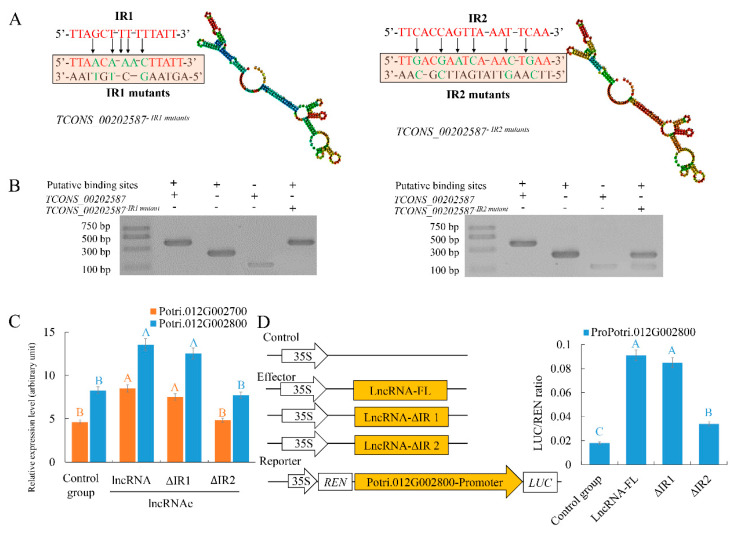
Figure 5.
Verification of the interaction of TCONS_00202587 with its target genes Potri.012G002700 and Potri.012G002800: (A) mutant sequence of putative interaction regions and (B) agarose gel electrophoresis of lncRNA and putative binding sites. Lane 1 was loaded with RNA containing the putative binding sites and synthetic TCONS_00202587 RNA. Lane 2 was loaded with putative binding sites. Lane 3 was loaded with synthetic TCONS_00202587 RNA. Lane 4 was loaded with putative binding sites and synthetic TCONS_00202587-I12-mutant/TCONS_00202587-IR2-mutant sequences. (C) Target expression patterns under LDH-NP binding with TCONS_00202587 and its putative interaction region mutant: control group represents non LDH-NPs binding with lncRNA. lncRNA-FL represents full-length lncRNA. ∆IR 1 and 2 represent deletion of putative interaction regions 1 and 2, respectively. (D) Transcriptional activation of TCONS_00202587 and its interaction region mutant on two targets: The data are mean ± SD of separate transfections (n = 3). Significantly different genes among the control group, LncRNA, ∆IR1, and ∆IR2 were determined using Fisher’s LSD test. Different colored letters on error bars indicate significant differences in the genes corresponding to that color among the groups at p < 0.01.