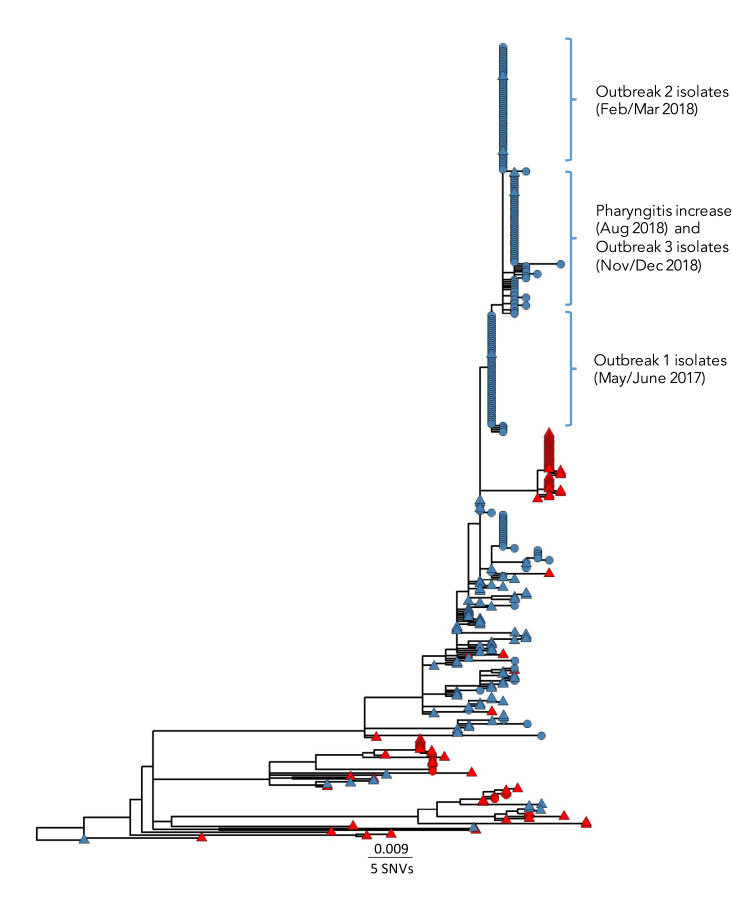
Figure 2.
Maximum likelihood core single nucleotide variant phylogenetic tree of S. pyogenes emm6.4 isolatesa collected in Canada, January 2012–January 2020 (n=403)
Abbreviation: SNV, single nucleotide variant
a A total of 529 sites were used in the phylogeny using 62% of the core genome. The scale bar represents the estimated evolutionary divergence between isolates based on the average genetic distance between strains (estimated number of sites of the isolate/total number of high quality single nucleotide variants). An internal isolate (sample number SC20-0734-A) was used as the mapping reference and the oldest outlying isolate (SC12-0215-A) was used as a root. Tip node colours represent isolates collected in Québec (blue) or other Canadian provinces (red), and tip node shapes represent non-invasive (circles) or invasive (triangles) Group A streptococcal infections