Abstract
American sloughgrass (Beckmannia syzigachne Steud.) is one of the most troublesome weeds infesting wheat and canola fields in China. Some biotypes cannot be controlled, either by acetolactate synthase (ALS) or acetyl coenzyme A carboxylase (ACCase) inhibitors, which are the main herbicides for controlling this weed. However, very few studies have investigated multiple resistance mechanism in B. syzigachne. In this study, a B. syzigachne biotype with a high resistance to ALS inhibitors we have reported was also showed relatively lower resistance to ACCase inhibitors, with a resistance index around 7. RNA-seq analysis was used to investigate the factors responsible for multiple resistance, and 60,108 unigenes were assembled by de novo transcriptome assembly and then annotated across eight databases. A Pro-197-Ser mutation was identified in the ALS gene by SNPs analysis and validated by PCR, while no mutation was identified in the ACCase gene. Nineteen candidate metabolic genes were screened and their overexpression was confirmed by qPCR. The expression of GST-T3 and GST-U6 in resistant plants ranged from 7.5- to 109.4-folds than that in susceptible ones at different times after two kinds of herbicide treatment. In addition, GST activities in resistant plants were 3.0–5.0 times higher than that in susceptible plants. Other novel resistance factors also showed high correlation with multiple resistance which included four genes encoding disease resistance proteins, a transcription factor (MYC3), and one gene conferring blight resistance. In this research, a B. syzigachne biotype was confirmed to have evolved multiple resistance to ACCase and ALS inhibitors. The Pro-197-Ser mutation in ALS gene and high-level GST activities were confirmed responsible for the multiple resistance. Characterized disease-resistance proteins, transcription factor, and blight-resistance proteins may play an essential role in these multiple herbicide resistance.
Keywords: herbicide resistance, weeds, metabolism, RNA-seq, overexpression
Introduction
American sloughgrass (Beckmannia syzigachne Steud.) is a troublesome diploid weed (2n = 14), which threatens many crop varieties in China such as wheat (Triticum aestivum L.) and oilseed rape [Brassica rapa L. subsp. Oleifera (DC.) Metzg.] (Rao et al., 2008; Li et al., 2015; Du et al., 2016; Li et al., 2017). Inhibitors of acetyl coenzyme A carboxylase (ACCase; EC 6.4.1.2) and acetolactate synthase (ALS; EC 4.1.3.18) are the main herbicides that are used to control the B. syzigachne in China. However, after many years of selection pressure, this weed evolved cross- or multiple-resistance to these two mechanisms of herbicides (Li et al., 2013; Li et al., 2015; Li et al., 2017). Resistant B. syzigachne can survive under high doses of these two kinds of herbicides via a variety of mechanisms. We sought to investigate resistance mechanisms useful for its control.
Based on the confirmed resistance against the ACCase-inhibiting and ALS-inhibiting herbicides in weeds, two kinds of resistance mechanisms can be summarized; these are target-site resistance (TSR) and non-target-site resistance (NTSR) (Délye, 2005). TSR is caused by amino acid substitutions in the conserved regions or due to differences in the expression of target enzyme genes, and has been well described (Délye, 2005; Powles and Yu, 2010). A mutation in the carboxyl-transferase (CT) domain of ACCase causes weed resistance to ACCase-inhibitors and confers distinct cross-resistance patterns (Powles and Yu, 2010; Scarabel et al., 2011; Beckie and Tardif, 2012; Kaundun, 2014). With respect to ALS, an amino acid substitution is commonly found at eight positions, and the different mutations also result in different levels of resistance against this kind herbicide (Yu and Powles, 2014b; Heap, 2019). NTSR is complicated and commonly results due to the overexpression of a set of genes that lead to reduced herbicide absorption, translocation, or increased metabolism and sequestration (Délye, 2013). To date, many well-established gene families are known to be involved in NTSR, such as the cytochrome P450 monooxygenase (P450s), and glutathione S-transferase (GSTs) families (Yuan et al., 2007). Many genes of these families are confirmed to have conferred resistance in herbicide-resistant weeds (Iwakami et al., 2014; Saika et al., 2014; Iwakami et al., 2019).
It has been reported that the mechanism governing the resistance of B. syzigachne to ACCase-inhibiting herbicides mainly involves TSR, in which amino acid substitutions have been identified at six positions (1781, 1999, 2027, 2041, 2078, and 2096) in resistant B. syzigachne (Du et al., 2016; Liu et al., 2019). Only the mutation involving Pro197Ser was found in the B. syzigachne population resistant to ALS-inhibiting herbicides (Li et al., 2015; Wang et al., 2018). For NTSR, metabolic genes related to resistance have been reported in fenoxaprop-P-ethyl-resistant B. syzigachne (Pan et al., 2016a; Pan et al., 2016b). However, to our knowledge, no studies have reported the involvement of genes in the NTSR mechanisms in B. syzigachne with ALS resistance, though the NTSR mechanisms have been confirmed (Wang et al., 2018; Wang et al., 2019; Bai et al., 2020). Furthermore, resistance genes relative to multiple resistance to ACCase and ALS-inhibiting herbicide have not been reported. Recently, we found a B. syzigachne biotype with ALS-inhibiting herbicide resistance which confirmed in our previous study was also hardly controlled by the ACCase inhibitors (Wang et al., 2018).
Hence, this research aimed to (1) determine the multiple resistance levels of B. syzigachne to ALS- and ACCase-inhibitors, (2) explore the multiple resistance factors using next generation sequencing methods, (3) confirm the Single Nucleotide Polymorphisms (SNPs) or the expression for the candidate genes, and (4) determine the enzyme activity which relative to the multiple resistance.
Materials and Methods
Plant Material and Whole Plant Assays
Resistant (R) B. syzigachne biotype were collected from Wuxi city (WC1148) and found evolved high-level resistance to ALS inhibitors in our previous study (Wang et al., 2018). A susceptible population (S) (WC1004), with no history of herbicide use was collected from near the R biotype. The plants of these two populations were cultured using the methods described in a previous study and thinned to 10 individuals per pot (Wang et al., 2018). Three kinds of ACCase inhibitor herbicide were selected for resistance assays (Table S1). The herbicides were applied when the seedlings reached the 3-leaf stage using a spray chamber with a moving TeeJet® XR8002 flat fan nozzle under a pressure of 0.275 MPa (Wang et al., 2018). Dry weights of the shoots (dried at 80°C for 48 h) in each pot were measured 21 days after treatment (DAT). The experiment of whole plant assay was repeated twice in a completely randomized design that had three biological replicates (three pots per treatment).
Sample Collection and RNA Extract for RNA-Seq
The plant culture conditions for the R and S populations were the same as in previous reports (Wang et al., 2018). The experimental design included three different treatment for R and S: without herbicide treatment control (CK), mesosulfuron-methyl treatment (M), and fenoxaprop-P-ethyl treatment (F). When the individuals reached the 3–5 leaf-stage, each biotype was treated with 70 g a.i. ha-1 of fenoxaprop-P-ethyl and the 20 g a.i. ha-1 mesosulfuron-methyl, respectively. After 24 h, the leaf tissues of three biological replicates were random collected, and also the samples without herbicide treatment were collected as controls with three individuals for each biotype. All the 18 samples were snap frozen in liquid nitrogen and stored at -80°C. Total RNA was extracted using the RNAprep Pure Plan Kit (Tiangen Biotech Beijing CO., LTD, Beijing, China). The degradation and contamination were checked by 1.0% agarose gel electrophoresis. RNA purity and concentration were determined using the NanoDrop-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). The RNA integrity was assessed by the Agilent Bio analyzer 2,100 system (Agilent Technologies, Palo Alto, USA) (Chen et al., 2017a).
Construction of cDNA Libraries, Sequencing, and Bioinformatics Analysis
The cDNA library construction was performed by a common method using the 18 detected RNA samples by Allgwegene Health Co. (Beijing, China) (Chen et al., 2017a). Paired-end reads (125 bp) were determined using an Illumina HiSeq™ 4000 (Illumina Inc., San Diego, CA, USA) platform, and clean reads were obtained from which raw reads with low quality were removed. Subsequently, de novo transcriptome assembly was carried out using the Trinity platform and the longest transcript of each locus was termed a “unigene” for subsequent annotation (Grabherr et al., 2011). Eight databases were selected for the annotation of the unigenes (Table S2). The software BLASTX (E-value < 10-5), BLAST2GO, and Blastall were used to predict and classify the different unigenes, as in previous studies (Tatusov et al., 2001; Ana et al., 2005; Clark et al., 2007).
SNPs Analysis for Target Genes and PCR Validation
For SNP calling, picard tools was used to map the paired-reads from each sample to the reference genome sequence of Brachypodium distachyon [L.] Beauv. The software GATK2 was used for SNP discovery and the detection stringency conditions include at least five reads calling the variant and >30 mapping quality score (Mckenna et al., 2010). To confirm the SNPs results for ALS and ACCase gene, the PCR method was selected to investigate the mutations in these two genes. DNA of the leaf tissues was exacted (15 individuals both for R and S populations) using the Hi-DNAsecure Plant Kit (DP350) by following the manufacturer’s instructions (Tiangen Biotech Beijing CO., Ltd., Beijing, China). Two pairs of PCR primers were design to clone ALS (1F- 5’CGCCTTACCCAAACCTACT3’, 1R- 5’ATGCGGCTGCTTGTTCTT3’; 2F- 5’ATCCCACCACAATATGCTATCC3’, 2R- 5’TCACAGTTGACCACACTTC3’); the amplification products of these two pairs of primers were 1035 and 766 bp in size, respectively, which encompass all of the reported mutation sites. One pair of primers was designed (F-5’AAACTCTGGTGCTCGGATTG3’; R-5’TAGGCTTCCATTTGCTCCC3’) to clone ACCase containing the CT region, and the PCR products were 1308 bp in size. The PCR mixture components and amplification conditions were in accordance with those reported in a previous study (Chen et al., 2017b), and the annealing temperature for all the primer pairs was 58°C. After confirming fragment amplification using 1% agarose gels prepared in 1× TAE, followed by staining with ethidium bromide, the PCR products were sequenced by Tsingke (Tsingke Biological Technology, Beijing Co., Ltd., Beijing, China), and the ALS and ACCase sequences of all the populations were aligned using the software DNAMAN (Lynnon BioSoft, San Ramon, CA, USA). The experiment of PCR was done twice and had 15 biological replicates both for R and S biotypes.
Analysis of Differentially Expressed Genes
Expression analysis was performed for the selected 18 samples, which were same as described above, and the samples were divided into different groups. The samples of the R and S biotypes without herbicide treatment were named CK1148 and CK1004, respectively. The sample groups treated with mesosulfuron-methyl for the R and S biotypes were named M1148 and M1004, respectively. Similarly, the samples groups treated with fenoxaprop-P-ethyl for the R and S populations were named F1148 and F1004, respectively. The cDNA library constructs and the clean reads detected for the 18 samples were the same as described above. The clean reads for the 18 libraries were mapped back onto the reference transcriptome, which was assembled in the above part by Bowtie 2 v.2.2.3 (Li and Dewey, 2011). The expression of each gene between sample pairs (CK1148 vs. CK1004, F1148 vs. CK1148, M1148 vs. CK1148, F1004 vs. CK1004, M1004 vs. CK1004, F1148 vs. F1004, and M1148 vs. M1004) were analyzed both by the numbers of reads and fragments aligned per thousand bases per million reads (FPKM), which was conducted using DESeq 2 v.1.4.5 (Audic and Claverie, 1997; Love et al., 2014). The false discovery rate is considered to be a standard to confirm the threshold of P values in multiple tests and analyses (Benjamini and Yekutieli, 2001).
Candidate Resistance Gene Selection and qPCR Validation
Candidate resistance genes were selected considering the reported NTSR genes include metabolism and signaling functions genes, the statistical significance (q < 0.05), and magnitude of differences in expression between the above treatment groups (|log2 (fold change)| ≥ 1). quantitative Real-time PCR (qPCR) was performed to confirm the accuracy of their expression. The best reference gene was selected from among capsine phosphatase (CAP), glyceraldehyde 3-phosphate dehydrogenase (CADP), and ubiquitin (UBQ) by the software BestKeeper (Pfaffl et al., 2004). The plant culture method, herbicide treatment for the individuals, and sample selection and storage were the same as described above. Total RNA was exacted, and first strand cDNA synthesis was performed (Tiangen Biotech Co., Ltd., Beijing, China) for all samples, and then primers were designed for all the genes (reference genes and the candidate resistance genes) using the software Oligo 7.0 (Table S3). The qPCR reaction was performed in 25 µL reaction volumes on an ABI 7500 PCR instrument under common conditions (Chen et al., 2017a). Relative expression of the selected genes was quantified using the 2-ΔΔCt method (Bustin et al., 2009). For the qPCR, three biological replicates were performed for each treatment, and three technical replicates was done for each sample.
Expression and Enzyme Activity Validation of GSTs
The plant culture for the R and S biotypes, and the herbicide treatment were same as described above. At 0, 24, 48, and 72 h, leaf samples for each treatment (at least three replicates) were collected and stored at -80°C. Total RNA was exacted and first strand cDNA synthesis was performed using the methods described above for all samples. The expression levels of two GSTs-related genes (GST-T3 and GST-U6) were detected using the qPCR method for all the samples, respectively, and the GST activity for the samples were also detected using a Glutathione S-transferase Microplate Assay Kit (CAK 1047) following the manufacturer’s instructions (Cohesion Biosciences Co., Ltd., Suzhou, China). Each sample (0.1 g leaf tissues) was assayed at 340 nm using a Tecan Infinite 200 Pro plate reader (Tecan Group Ltd., Männedorf, Switzerland). The results were expressed as U g-1, and one unit of GST activity was defined as the enzyme generating 1 μmol of glutathione and 1-chloro-2, 4-dinitrobenzene conjugate in the reaction time. Both the gene expression and GST activity assay were repeated twice and at least three biological replicates were performed for each treatment.
Statistical Analysis
The values of herbicide doses required to reduce 50% of the plant growth (GR50) were estimated through nonlinear regression with a log-logistic model:
using Sigma Plot 12.0 (Systat software, San Jose, CA, USA) (Seefeldt et al., 1995). In this model, b is the slope of the curve, y0 is the lower limit, a is the difference between the upper limit and the lower limit, and X0 is the GR50. Resistance levels were determined by calculating the ratio of the GR50 of the resistant biotype to that of the susceptible biotype, and the results are expressed as resistance index (RI). The difference of gene expression and GST activity between R and S biotypes were analyzed by the Student’s t-test, which were performed with SPSS 13.0 (SPSS, Chicago, USA).
Results
Resistance Levels to ALS- and ACCase-Inhibiting Herbicides
The sensitivity of the R (WC1148) and S (WC1004) biotypes in response to four different ACCase inhibitors (fenoxaprop-P-ethyl, clodinafop-propargyl, clethodim, and pinoxaden) was calculated. The GR50 values of the S biotype WC1004 were 42.4, 12.1, 11.4, and 11.8 g a. i. ha-1, respectively. For the R biotype WC1148, the GR50 values were 87.0, 39.4, 84.5, and 89.2 g a. i. ha-1, respectively. Compared with those of the sensitive biotypes, the RI of WC1148 were 2.1, 3.3, 7.4 and 7.5, respectively (Table 1). These results indicated that the ALS inhibitors resistant biotype WC1148 evolved multiple resistance to the ACCase inhibitors.
Table 1.
GR50 values and resistance index of B. syzigachne to ACCase inhibitors in the R and S biotypes.
| Classification of herbicidea | Herbicides | Biotypesb | GR50 ± SE (g a.i.ha-1) | P-value | Resistanceindex (RI)c | |
|---|---|---|---|---|---|---|
| ACCase inhibitors | APPs | Fenoxaprop-P-ethyl | S | 42.4 ± 9.8 | <0.001 | — |
| R | 87.0 ± 24.8 | <0.001 | 2.1 | |||
| Clodinafop-propargyl | S | 12.1 ± 9.2 | <0.001 | — | ||
| R | 39.4 ± 7.8 | <0.001 | 3.3 | |||
| CHDs | Clethodim | S | 11.4 ± 5.7 | <0.001 | — | |
| R | 84.5 ± 45.5 | <0.001 | 7.4 | |||
| PPZs | Pinoxaden | S | 11.8 ± 3.6 | <0.001 | — | |
| R | 89.2 ± 65.9 | <0.001 | 7.5 | |||
APPs, Aryloxyphenoxypropionates; CHDs, Cyclohexanediones; PPZs, Phenylpyrazoline;
S: susceptible biotype; R: resistant biotype; GR50: herbicide doses required to inhibit dry weight by 50% compared to untreated controls;
RI, resistance index, and determined by dividing the GR50 of the R by that of S biotype.
RNA-Seq and De Novo Assembly, Gene Annotation, and Functional Classification
The OD260/OD280 values (around 2.0) and the RNA integrity number (RIN) values (>6.0) indicated that the RNA integrity of the samples was suitable for the requirements of subsequent experiments. The cDNA library of 18 RNA samples were analyzed: 449,583,465 raw reads, and 436,188,739 clean reads which ranged from 22,724,569 to 24,516,824 per sample were generated, respectively (Table S1). We obtained 118,111 unigenes that ranged from 201 to 21,671 bp in size and with an N50 of 1,328 bp.
More than half (50.89%) of the total 118,111 unigenes were successfully annotated in at least one database, and 8,197 unigenes were annotated in all of the database (Table S1). 35,616 unigenes were assigned into 42 functional categories by the Gene Ontology (GO) database and including 20 for “biological process”, 12 for “cellular component”, and 10 for “molecular function” (Table S4). In total, 25,598 unigenes were categorized into 26 Clusters of Orthologous Groups of proteins (KOG) classifications (Table S5). The 7,893 assembled sequences were mapped to the reference canonical pathway in Kyoto Encyclopedia of Genes and Genomes (KEGG) (Table S6). The 48,600 unigenes were assigned putative annotations by the NCBI non-redundant protein sequences (NR), 39,869 unigenes by NCBI non-redundant nucleotide sequences (NT), 31,291 unigenes by a manually annotated and reviewed protein sequence database (SwissProt), and 39,310 unigenes in the Protein family (Pfam) database.
SNPs Analysis and PCR Validation for ALS and ACCase Gene
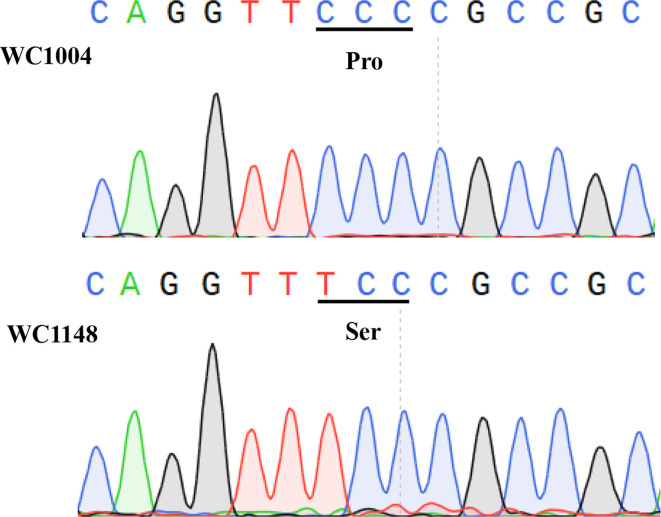
Two SNPs were detected for the ALS gene (c38811_g1) using the RNA-seq analysis, while, no SNPs was found for the ACCase gene (Table S7). For the ALS gene, one SNP was only found in the resistant individuals (C to T at position 629) which causing the amino acid Pro change to Ser (Table S7). While, another SNP (G to A at position 820) was only found in the susceptible individuals but without causing any amino acid change. To confirm the results of the SNPs assay, parts of the ALS and ACCase genes which contain all the common mutation sites of these two kinds of inhibitors in B. syzigachne were cloned from the R and S populations. Comparing the conserved regions of the ALS gene for the S and R biotypes, it was found that 197 sites of ALS were mutated from proline CCC (Pro) to serine TCC (Ser), and no other mutation sites were found in any of the 15 individuals (Figure 1). By comparing the ACCase CT region of the R and S biotypes, no mutation was found in the CT region of the ACCase gene. The results of the chromatograms show that the ALS gene in the R biotypes was a single peak at the mutation site, indicating a homozygous mutation (Figure 1), and the detected 15 individuals of R biotypes showed the same mutation type.
Figure 1.
Sequences result for ALS genes in WC1148 and WC1004 biotypes, respectively. A Pro-107-Ser mutation in the ALS gene evolved in the resistant biotype WC1148, and DNA sequencing chromatograms show the homozygous mutation.
Analysis of Differentially Expressed Genes
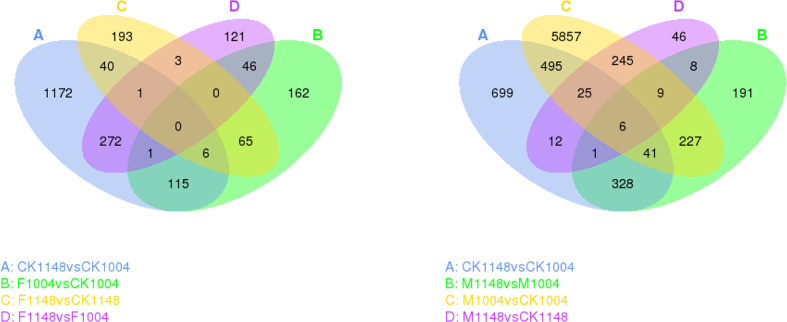
Seven different comparison combinations were designed as the biotype WC1148 showed multiple resistance to ALS and ACCase inhibitor herbicides (Figure 2). Differentially expressed genes between these groups (|log2 (fold change)| ≥ 1 and q < 0.05) were selected as candidate resistance genes. The number of up- and down-regulated genes in CK1148 vs CK1004 combination was 700 and 907, respectively. For the group set of M1148 vs CK1148, and F1148 vs CK1148, the number of up- and down-regulated genes was no more than 300. After herbicide treatment, differentially expressed genes between the R and S biotypes (M1148 vs. M1004 and F1148 vs. F1004) were more than 400. The number of up- and down-regulated genes in the M1004 vs. CK1004 combination was 2,720 and 4,185, respectively. However, the number of up- and down-regulated genes in the F1004 vs. CK1004 combination was 162 and 233, respectively (Table 2).
Figure 2.
Venn diagram showing differential gene expression between the different group sets for the resistant biotype WC1148 and susceptible biotype WC1004. The circle represents the comparisons, the sum of the numbers in each circle represents the genes showing differential expression, and the overlapping part of the circle represents the number of the same genes between different comparisons.
Table 2.
Number of differentially expressed genes among different groups.
| Treatment groups | Differentially expressed gene | Up-regulated | Down-regulated |
|---|---|---|---|
| CK1148 vs. CK1004 | 1607 | 700 | 907 |
| M1148 vs. CK1148 | 352 | 274 | 78 |
| F1148 vs. CK1148 | 308 | 115 | 193 |
| M1004 vs. CK1004 | 6905 | 2720 | 4185 |
| F1004 vs. CK1004 | 395 | 162 | 233 |
| M1148 vs. M1004 | 811 | 393 | 418 |
| F1148 vs. F1004 | 444 | 203 | 241 |
CK: untreated herbicides; M: mesosulfurn-methyl treatment; F: fenoxaprop-p-ethyl treatment.
Selection of Candidate Non-Target Site Resistance Genes
We analyzed and selected factors related to NTSR based on the reported mechanism in weeds (Délye, 2013; Yu and Powles, 2014a; Pan et al., 2016a). The reported genes relative to the metabolic enzyme families in B. syzigachne were considered which include CytP450, GST, ABC transporter family, and esterase gene families (Pan et al., 2016a). Other genes up-regulated in all the comparative groups were considered, especially for up-regulated genes in CK1148 vs. CK1004. After preliminary analysis of the different expression of the genes between different treatment groups, 19 candidate genes were related to resistance. Among these candidate genes, common metabolic genes were identified, which included three genes annotated as GSTs, one gene being an ABC transporter, two protein kinase genes, one annotation as UDP-glycosyl transferase, and two for oxidases. In addition, certain novel resistant factors were found, which included two immune proteins, five disease resistance proteins, two termed ‘other’ transferase, and one transcription factor (Table 3).
Table 3.
Candidate genes related to mesosulfuron-methyl and fenoxaprop-p-ethyl metabolism in B. syzigachne.
| Gene ID | Gene annotation | Fold-change(F1148 vs F1004 or M1148 vs M1004) | Padj |
|---|---|---|---|
| c59327_g1 | GST-T3 | 8.68 | 2.22 × 10-20 |
| c9871_g1 | GST-U6 | 4.82 | 2.62 × 10-20 |
| c81158_g1 | GST-U6 | 5.46 | 2.05 × 10-3 |
| c61256_g2 | ABC G family member 15 | 5.81 | 3.57 × 10-20 |
| c52253_g1 | UDP-glycosyltransferase 88F5 | 6.57 | 6.51 × 10-20 |
| c30288_g1 | LRR RLK At4g08850 | 8.36 | 1.60 × 10-9 |
| c64517_g5 | LRR RLK At1g56140 | 3.95 | 8.94 × 10-3 |
| c61823_g3 | Iron/ascorbate family oxidoreductases | 6.82 | 9.95 × 10-7 |
| c50888_g2 | L-pipecolate oxidase | 4.03 | 3.25 × 10-3 |
| c61143_g4 | Late blight resistance protein R1B-16 | 10.86 | 6.83 × 10-20 |
| c71182_g1 | Late blight resistance protein R1A-10 | 7.76 | 2.59 × 10-4 |
| c41866_g1 | Disease resistance protein At1g15890 | 10.52 | 7.67 × 10-13 |
| c62003_g3 | Disease resistance protein At1g58602 | 9.99 | 4.94 × 10-10 |
| c63887_g5 | Disease resistance protein RPP13 | 5.33 | 1.10 × 10-6 |
| c64184_g2 | Disease resistance protein RGA3 | 4.30 | 1.15 × 10-4 |
| c51431_g2 | Disease resistance protein RPP13 | 7.11 | 5.85 × 10-4 |
| c59149_g1 | Polyamine aminopropyltransferase | 5.99 | 1.40 × 10-10 |
| c45597_g1 | Anthranilate N-benzoyltransferase protein 1 | 6.79 | 6.83 × 10-4 |
| c96180_g1 | Transcription factor MYC3 | 3.77 | 4.11 × 10-2 |
qPCR Validation of Candidate Metabolic Resistance Gene Expression
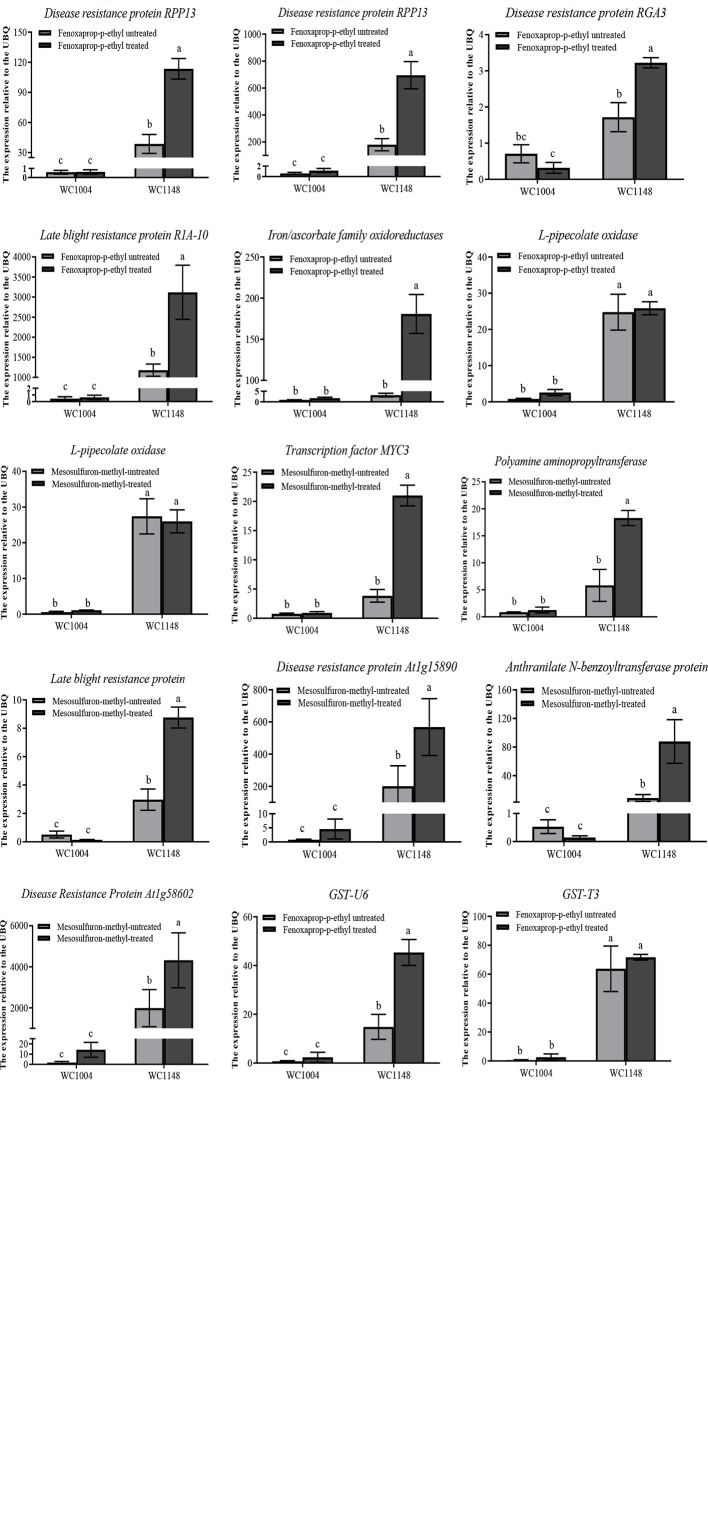
The expression of the selected contigs were confirmed using the qPCR method, and UBQ was the best reference gene which analyzed by Bestkeeper (Table S8) (Pfaffl et al., 2004). The expression levels of 13 candidate resistance genes were validated by qPCR at 0 and 24 h after treatment with 20 g a.i. ha-1 mesosulfuron-methyl for sensitive R and S biotypes. The results showed that in the absence of treatment with the ALS inhibitor mesosulfuron-methyl, seven genes, including GST-U6, disease resistance protein At1g4809, and disease resistance protein At1g58062 in the R biotype, exhibited 5.8-1124.0 times higher expression than that in S (Figure 3). However, other genes show similar expression level between the R and S biotypes (Figure 3). After this herbicide treatment, all these candidate genes showed 5.9–4928.2 times higher expression levels in the R biotype than that in the S biotype.
Figure 3.
Relative expression validations for candidate resistance genes using the qPCR method. The confirmed reference gene UBQ was selected as an internal control gene. The resistant biotype named WC1148 and the susceptible biotype named WC1004. Three biological replicates were performed for each treatment, and three technical replicates was done for each sample. Vertical bars show the standard error of the mean for the replicates. Different lowercase characters indicate significant differences in expression between the treatments.
Moreover, eight candidate resistance genes were validated by qPCR at 0 and 24 h after treatment with 70 g a.i. ha-1 fenoxaprop-p-ethyl. Among these genes, six showed higher expression in the R biotype both before and after herbicide treatment than that in S biotype. In particular, for the gene disease resistance protein RPP13, without herbicide treatment, the expression level in the R biotype was 300 times higher than in the S biotype (Figure 3). After herbicide treatment, the expression could reach 700 times higher.
Expression and Enzyme Activity Validation of GSTs
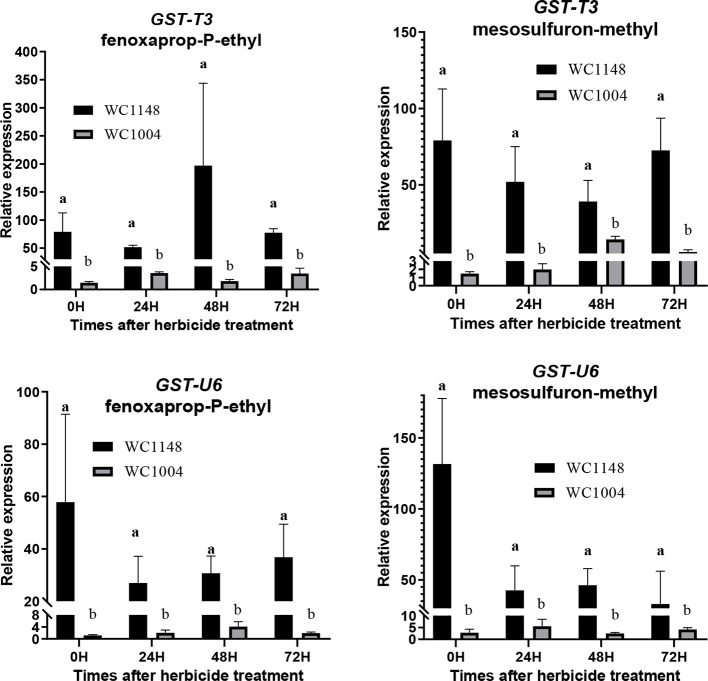
The expression levels of two unigenes (GST-T3, GST-U6) annotation to GSTs were analyzed at different times after herbicide treatment. The results show that expression of these two genes in the R individuals was higher than that in the S individuals at each time point under herbicide treatment (Figure 4). After mesosulfuron-methyl treatment, the expression of GST-T3 in R can reach to 53.8-folds than S, similar results (108.5-folds) were also found after fenoxaprop-P-ethyl treatment. Without herbicide treatment, the expression of GST-U6 was 46.5 times higher than in S and show slight decreased after these two kinds herbicide treatment.
Figure 4.
The expression of two unigenes (GST-T3, GST-U6) annotation to GSTs in the resistant (R) and susceptible (S) plants at 0, 24, 48, and 72 h after mesosulfuron-methyl (20 g a.i. ha-1) and fenoxaprop-P-ethyl (70 g a.i. ha-1) treatment, respectively. At least three biological replicates were performed for each treatment, and three technical replicates was done for each sample. Bars are means ± standard error of the mean (SEM). Different lowercase means the significant difference at each time point between R and S which analyzed by the T-test (P < 0.05).
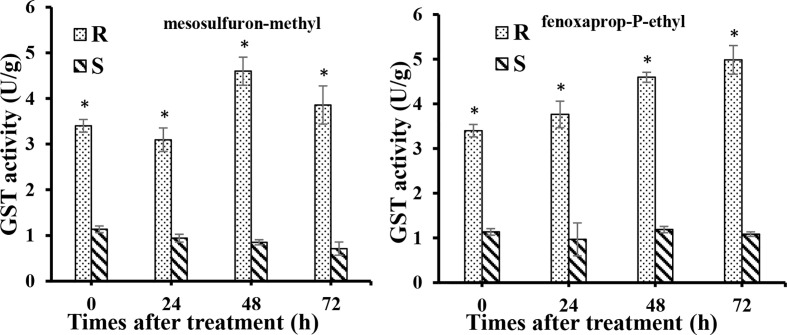
To determine whether the higher expression of these genes of GSTs increase the GST active in R biotype. GST activity was investigated in both R and S biotypes at 0, 24, 48, and 72 h after mesosulfuron-methyl and fenoxaprop-P-ethyl treatment. The results show that in R plants without herbicide treatment, the GST activity was 3.4 U g-1, which was 3.0 times higher than that in the S plants (Figure 5). After mesosulfuron-methyl treatment, GST activities were slightly increased in R biotype and reached a peak at 48 h (4.6 U g-1), which was 5.0 times higher than that in S biotype. However, it was slightly decreased in the S biotype, which is around 0.8 U g-1. Similar results were also found in these two biotypes after fenoxaprop-P-ethyl treatment. The GST activities were increased in R biotype and can reach to 4.9 U g-1, while GST activities in the S biotype was around 1.0 U g-1 at all the time points. GST activities in the R plants were significantly higher than those in the S plants at all the time points.
Figure 5.
GST activities in the resistant (R) and susceptible (S) plants at 0, 24, 48, and 72 h after fenoxaprop-P-ethyl (70 g a.i. ha-1) and mesosulfuron-methyl (20 g a.i. ha-1) treatment, respectively. At least three biological replicates were performed for each treatment. Bars are means ± standard error of the mean (SEM). *Significant difference at each time point between R and S (P < 0.05).
Discussion
Multiple Resistance Levels to ALS and ACCase Inhibitors
In this study, we confirmed that a B. syzigachne biotype had evolved multiple resistance to ALS and ACCase inhibitor herbicides. In our previous research, this biotype showed higher resistance to mesosulfuron-methyl, and also exhibited higher cross-resistance to other ALS inhibitor herbicides, such as flucarbazone, imazapic, pyroxsulam, and pyribenzoxim (Wang et al., 2018). In this study, the same biotype shows relatively low resistance levels to the ACCase inhibitor herbicide, with resistance levels ranging from 2.1 to 7.5. Cross-resistance is a common phenomenon in weed species resistant to ALS and ACCase inhibitor herbicides, such as Conyza canadensis (Zheng et al., 2011), Myosoton aquaticum L. (Liu et al., 2015), and Bromus tectorum L. (Park and Mallory-Smith, 2004). Beckmannia syzigachne, biotypes with cross-resistance to ACCase and ALS inhibitor herbicides are frequently reported in China (Li et al., 2013; Li et al., 2014; Li et al., 2015; Pan et al., 2015; Tang et al., 2015; Du et al., 2016; Liu et al., 2019; Wang et al., 2019). In addition, a biotype was also reported to have evolved multiple resistance to these kinds of herbicide (Li et al., 2017). In contrast to the biotypes described in this research, this reported biotype showed low resistance to the ALS inhibitor mesosulfuron-methyl with an RI of 2.6. Furthermore, this biotype showed high resistance to the ACCase inhibitor fenoxaprop-P-ethyl, with an RI value of 31.2 (Li et al., 2017).
Target-Site Resistance Mechanisms in This Biotype
Mutations involving the target gene ALS and ACCase is a common mechanism for ALS and ACCase inhibitor herbicide resistance. In this research, the ALS gene in the R biotype was shown to have a Pro-197-Ser mutation both by the SNPs and PCR analyses, this indicate the mutation may be one of a number of resistance mechanisms. Evolved resistance in weeds has been attributed to substitutions at each of the following eight different amino acids: Ala-122, Pro-197, Ala-205, Asp-376, Arg-377, Trp-574, Ser-653, and Gly-654 (Murphy and Tranel, 2019). Mutation at the site of 197 of the ALS gene was the most frequently reported mutation type in weed species, and was first found in Kochia scoparia (L.) Schrad (Saari et al., 1990). For B. syzigachne, only one kind of mutation (Pro-197-Ser) was found among the biotypes resistant to ALS inhibitor (Li et al., 2015). To date, seven kinds of amino acid substitutions in the CT domain of ACCase have been reported as being related to resistance to ACCase inhibitor herbicides in grassy weeds, and six substitutions have been found in B. syzigachne. In this research, no mutations were found in the R biotype, this indicated NTSR may be evolved in this biotype.
Common Metabolic Genes Relative to Multiple Resistance
Herbicide metabolism in weeds is a complex process involving metabolic enzymes (P450, GST, esterase, transporter, oxidase) and regulatory genes (transcription factors, protein kinases, and micro-RNAs) (Kreuz et al., 1996; Van Eerd et al., 2003; Yuan et al., 2007). Beckmannia syzigachne has evolved resistance to ACCase inhibitors, and many genes involved in NTSR were discovered using the RNA-seq technology (Pan et al., 2016a). Metabolic genes including CytP450, GST, esterases, peroxidases, ABC transporter B family members, and UDP-glycosyltransferases were identified (Pan et al., 2016a) Esterases break down herbicide molecules, and ABC transporters enhance resistance to herbicides by transporting isolated herbicides and acting as metabolites (Délye, 2013). In this research, one gene was annotated as an ABC transporter G family member, and one gene was annotated as UDP-glycosyltransferase 88F5 may be associated with resistance. Furthermore, two genes (GST-T3 and GST-U6) were found to be overexpressed at different times after mesosulfuron-methly and fenoxaprop-p-ethyl treatment in resistant B. syzigachne. What is more, the GST activity in resistant plants higher than the susceptible ones at all the detected times. This result is similar to that reported in a previous study (Pan et al., 2016b). This confirmed that these two genes are related to multiple resistance in this weed.
Novel Genes Related to Multiple Resistance
In addition to the reported resistance factors in B. syzigachne, some new genes related to resistance were also found in this research. Four genes annotated as encoding disease resistance proteins (one for At1g15890, one for At1g58602, and two for RPP13) in the resistant population were expressed at 65.8-1124.0 times higher levels than that in the sensitive biotype. This phenomenon was found both before and after herbicide treatment. This suggested that the disease-resistance protein might be related to the resistance. The expression levels of oxidase (L-pipecolate oxidase and iron/ascorbate family oxidoreductases) in R biotypes were 10.0–103.4 times higher than those in sensitive biotypes after treatment, which may be related to fenoxaprop-p-ethyl metabolism in individuals.
In this study, besides disease-resistant proteins, transcription factors and other regulatory genes were also screened. Protein kinases play key roles in signal transduction pathways, bringing external signals into cells, amplifying cascades of reactions, and passing them to transcription factor proteins to regulate the expression of downstream functional genes. Plant immune receptor is often classified as threonine kinase, and studies have shown that tyrosine phosphorylation in plants has an important regulatory role in innate immunity (Liu et al., 2018). MYC2 has a regulatory role in plant antioxidant capacity (Dombrecht et al., 2007). In this study, MYC3 transcription factors and protein kinase levels in WC11-48 expression were 22.8-917.2 times higher in the resistant biotype than that in the sensitive biotype, but the present study has not reported anything about the relationship between resistance genes and weeds. The roles of these genes in weed resistance mechanisms still need further research.
Conclusion
In this research, we found a B. syzigachne biotype that had evolved multiple resistance to ALS and ACCase inhibitor herbicides, and which showed high resistance to ALS inhibitor and relatively lower resistance to the ACCase ones. A Pro-197-Ser mutation was found in the ALS gene and no mutations were found in the ACCase gene. Four genes encoding disease resistance proteins, the MYC3 transcription factor, and one gene for blight resistance protein were found related to confer multiple resistance. Of the novel resistance factors, two GST genes (GST-T3 and GST-U6) showed high correlation to this multiple resistance. The results of this research could provide the important resistant factors about multiple resistance to ACCase and ALS-inhibiting herbicide in B. syzigachne.
Data Availability Statement
The raw Illumina sequence reads have been deposited in the NCBI Sequence Read Archive (SRA) database. The accession numbers for the samples with herbicide treatment were SRR10737896 and SRR10737895. And the accession numbers for the samples without herbicide treatment were SRR10742660 and SRR10742659.
Author Contributions
This research was designed by HC. JW and JC performed the experimental work and the data analysis. XL, HC, DL, ZL, and JC provided helpful suggestions for the data analysis and manuscript revision. JC wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by The National Natural Science Foundation of China (no. 31371952).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are thankful to The National Natural Science Foundation of China for financial support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.572610/full#supplementary-material
References
- Ana C., Stefan G. T., Juan Miguel G. G., Javier T., Manuel T., Montserrat R. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21 (18), 3674–3676. 10.1093/bioinformatics/bti610 [DOI] [PubMed] [Google Scholar]
- Audic S., Claverie J. M. (1997). The significance of digital gene expression profiles. Genome Res. 7 (10), 986–995. 10.1101/gr.7.10.986 [DOI] [PubMed] [Google Scholar]
- Bai S., Zhao Y., Zhou Y., Wang M., Li Y., Luo X., et al. (2020). Identification and expression of main genes involved in non-target site resistance mechanisms to fenoxaprop-p-ethyl in Beckmannia syzigachne. Pest Manag. Sci. 76 (8), 2619–2626. 10.1002/ps.5800 [DOI] [PubMed] [Google Scholar]
- Beckie H. J., Tardif F. J. (2012). Herbicide cross resistance in weeds. Crop Prot. 35, 15–28. 10.1016/j.cropro.2011.12.018 [DOI] [Google Scholar]
- Benjamini Y., Yekutieli D. (2001). The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 29 (4), 1165–1188. 10.1214/aos/1013699998 [DOI] [Google Scholar]
- Bustin S. A., Benes V., Garson J. A., Hellemans J., Huggett J., Kubista M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55 (4), 611–622. 10.1373/clinchem.2008.112797 [DOI] [PubMed] [Google Scholar]
- Chen J., Huang H., Wei S., Huang Z., Wang X., Zhang C. (2017. a). Investigating the mechanisms of glyphosate resistance in goosegrass (Eleusine indica (L.) Gaertn.) by RNA sequencing technology. Plant J. 89 (2), 407–415. 10.1111/tpj.13395 [DOI] [PubMed] [Google Scholar]
- Chen J., Jiang C., Huang H., Wei S., Huang Z., Wang H., et al. (2017. b). Characterization of Eleusine indica with gene mutation or amplification in EPSPS to glyphosate. Pestic. Biochem. Phys. 143, 201–206. 10.1016/j.pestbp.2017.09.012 [DOI] [PubMed] [Google Scholar]
- Clark R. M., Gabriele S., Christopher T., Stephan O., Georg Z., Paul S., et al. (2007). Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science 317 (2007), 338–342. 10.1126/science.1138632 [DOI] [PubMed] [Google Scholar]
- Délye C. (2005). Weed resistance to acetyl coenzyme A carboxylase inhibitors: an update. Weed Sci. 53 (5), 728–746. 10.1614/WS-04-203R.1 [DOI] [Google Scholar]
- Délye C. (2013). Unravelling the genetic bases of non-target-site-based resistance (NTSR) to herbicides: a major challenge for weed science in the forthcoming decade. Pest Manag. Sci. 69 (2), 176–187. 10.1002/ps.3318 [DOI] [PubMed] [Google Scholar]
- Dombrecht B., Xue G. P., Sprague S. J., Kirkegaard J. A., Ross J. J., Reid J. B., et al. (2007). MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19 (7), 2225–2245. 10.1105/tpc.106.048017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Liu W., Yuan G., Guo W., Li Q., Wang J. (2016). Cross-resistance patterns to ACCase-inhibitors in American sloughgrass (Beckmannia syzigachne Steud.) homozygous for specific ACCase mutations. Pestici. Biochem. Phys. 126, 42–48. 10.1016/j.pestbp.2015.07.005 [DOI] [PubMed] [Google Scholar]
- Grabherr M. G., Haas B. J., Moran Y., Levin J. Z., Thompson D. A., Ido A., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29 (7), 644–652. 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heap I. (2019). The international survey of herbicide resistant weeds. Available at: http://www.weedscience.org/ (Accessed December 15, 2019).
- Iwakami S., Uchino A., Kataoka Y., Shibaike H., Watanabe H., Inamura T. (2014). Cytochrome P450 genes induced by bispyribac-sodium treatment in a multiple-herbicide-resistant biotype of Echinochloa phyllopogon. Pest Manag. Sci. 70 (4), 549–558. 10.1002/ps.3572 [DOI] [PubMed] [Google Scholar]
- Iwakami S., Kamidate Y., Yamaguchi T., Ishizaka M., Endo M., Suda H., et al. (2019). CYP 81A P450s are involved in concomitant cross-resistance to acetolactate synthase and acetyl-CoA carboxylase herbicides in Echinochloa phyllopogon. New Phytol. 221 (4), 2112–2122. 10.1111/nph.15552 [DOI] [PubMed] [Google Scholar]
- Kaundun S. S. (2014). Resistance to acetyl-CoA carboxylase-inhibiting herbicides. Pest Manag. Sci. 70 (9), 1405–1417. 10.1002/ps.3790 [DOI] [PubMed] [Google Scholar]
- Kreuz K., Tommasini R., Martinoia E. (1996). Old enzymes for a new job (herbicide detoxification in plants). Plant Physiol. 111 (2), 349–353. 10.1104/pp.111.2.349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Dewey C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 12 (1):1. 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Bi Y., Liu W., Yuan G., Wang J. (2013). Molecular basis for resistance to fenoxaprop-p-ethyl in American sloughgrass (Beckmannia syzigachne Steud.). Pestic. Biochem. Phys. 105 (2), 118–121. 10.1016/j.pestbp.2012.12.007 [DOI] [Google Scholar]
- Li L., Du L., Liu W., Yuan G., Wang J. (2014). Target-site mechanism of ACCase-inhibitors resistance in American sloughgrass (Beckmannia syzigachne Steud.) from China. Pestic. Biochem. Phys. 110 (5), 57–62. 10.1016/j.pestbp.2014.03.001 [DOI] [PubMed] [Google Scholar]
- Li L., Liu W., Chi Y., Guo W., Luo X., Wang J. (2015). Molecular mechanism of mesosulfuron-methyl resistance in multiply-resistant American sloughgrass (Beckmannia syzigachne). Weed Sci. 63 (4), 781–787. 10.1614/WS-D-15-00026.1 [DOI] [Google Scholar]
- Li W., Zhang L., Zhao N., Guo W., Liu W., Li L., et al. (2017). Multiple resistance to ACCase and ALS-inhibiting herbicides in Beckmannia syzigachne (Steud.) Fernald without mutations in the target enzymes. Chil. J. Agr. Res. 77 (3), 257–265. 10.4067/S0718-58392017000300257 [DOI] [Google Scholar]
- Liu W., Yuan G., Du L., Guo W., Li L., Bi Y., et al. (2015). A novel Pro197Glu substitution in acetolactate synthase (ALS) confers broad-spectrum resistance across ALS inhibitors. Pestic. Biochem. Phys. 117, 31–38. 10.1016/j.pestbp.2014.10.005 [DOI] [PubMed] [Google Scholar]
- Liu J., Liu B., Chen S., Gong B., Chen L., Zhou Q., et al. (2018). A tyrosine phosphorylation cycle regulates fungal activation of a plant receptor Ser/Thr kinase. Cell Host. Microbe 23 (2), 241–253. e246. 10.1016/j.chom.2017.12.005 [DOI] [PubMed] [Google Scholar]
- Liu B., Ding F., Wang M., Wang F., Luo X., Li L. (2019). Cross-resistance pattern to ACCase-inhibiting herbicides in a novel Trp1999Leu mutation American sloughgrass (Beckmannia syzigachne) population. Pestic. Biochem. Phys. 159, 80–84. 10.1016/j.pestbp.2019.05.019 [DOI] [PubMed] [Google Scholar]
- Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Bio 15 (12):550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mckenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., et al. (2010). The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20 (9), 1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy B. P., Tranel P. J. (2019). Target-Site Mutations Conferring Herbicide Resistance. Plants 8 (10), 382. 10.3390/plants8100382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L., Li J., Zhang T., Zhang D., Dong L. (2015). Cross-resistance patterns to acetyl coenzyme A carboxylase (ACCase) inhibitors associated with different ACCase mutations in Beckmannia syzigachne. Weed Res. 55 (6), 609–620. 10.1111/wre.12170 [DOI] [Google Scholar]
- Pan L., Gao H., Xia W., Zhang T., Dong L. (2016. a). Establishing a herbicide-metabolizing enzyme library in Beckmannia syzigachne to identify genes associated with metabolic resistance. J. Exp. Bot. 67 (6), 1745–1757. 10.1093/jxb/erv565 [DOI] [PubMed] [Google Scholar]
- Pan L., Wang Z., Cai J., Gao H., Zhao H., Dong L. (2016. b). High-throughput sequencing reveals differential regulation of miRNAs in fenoxaprop-P-ethyl-resistant Beckmannia syzigachne. Sci. Rep. 6, 28725. 10.1038/srep28725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park K., Mallory-Smith C. (2004). Physiological and molecular basis for ALS inhibitor resistance in Bromus tectorum biotypes. Weed Res. 44 (2), 71–77. 10.1111/j.1365-3180.2003.00374.x [DOI] [Google Scholar]
- Pfaffl M. W., Tichopad A., Prgomet C., Neuvians T. P. (2004). Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol. Lett. 26 (6), 509–515. 10.1023/b:bile.0000019559.84305.47 [DOI] [PubMed] [Google Scholar]
- Powles S. B., Yu Q. (2010). Evolution in action: plants resistant to herbicides. Annu. Rev. Plant Biol. 61, 317–347. 10.1146/annurev-arplant-042809-112119 [DOI] [PubMed] [Google Scholar]
- Rao N., Dong L., Li J., Zhang H. (2008). Influence of environmental factors on seed germination and seedling emergence of American sloughgrass (Beckmannia syzigachne). Weed Sci. 56 (4), 529–533. 10.1614/WS-07-158.1 [DOI] [Google Scholar]
- Saari L. L., Cotterman J. C., Primiani M. M. (1990). Mechanism of sulfonylurea herbicide resistance in the broadleaf weed, Kochia scoparia. Plant Physiol. 93 (1), 55–61. 10.1104/pp.93.1.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saika H., Horita J., Taguchi-Shiobara F., Nonaka S., Nishizawa-Yokoi A., Iwakami S., et al. (2014). A novel rice cytochrome P450 gene, CYP72A31, confers tolerance to acetolactate synthase-inhibiting herbicides in rice and Arabidopsis. Plant Physiol. 166 (3), 1232–1240. 10.1104/pp.113.231266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarabel L., Panozzo S., Varotto S., Sattin M. (2011). Allelic variation of the ACCase gene and response to ACCase-inhibiting herbicides in pinoxaden-resistant Lolium spp. Pest Manag. Sci. 67 (8), 932–941. 10.1002/ps.2133 [DOI] [PubMed] [Google Scholar]
- Seefeldt S. S., Jensen J. E., Fuerst E. P. (1995). Log-logistic analysis of herbicide dose-response relationships. Weed Technol. 9 (2), 218–227. 10.1017/S0890037X00023253 [DOI] [Google Scholar]
- Tang W., Zhou F., Zhang Y., Chen J. (2015). Resistance of American sloughgrass (Bechmannia syzigachne) populations to ACCase-inhibiting herbicides involves three different target site mutations from China. Pestic. Biochem. Phys. 124, 93–96. 10.1016/j.pestbp.2015.05.003 [DOI] [PubMed] [Google Scholar]
- Tatusov R. L., Natale D. A., Garkavtsev I. V., Tatusova T. A., Shankavaram U. T., Rao B. S., et al. (2001). The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 29 (1), 22–28. 10.1093/nar/29.1.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Eerd L. L., Hoagland R. E., Zablotowicz R. M., Hall J. C. (2003). Pesticide metabolism in plants and microorganisms. Weed Sci. 51 (4), 472–495. 10.1614/0043-1745(2003)051[0472:PMIPAM]2.0.CO;2 [DOI] [Google Scholar]
- Wang J., Li X., Li D., Han Y., Li Z., Yu H., et al. (2018). Non-target-site and target-site resistance to AHAS inhibitors in American sloughgrass (Beckmannia syzigachne). J. Integr. Agr. 17 (12), 2714–2723. 10.1016/S2095-3119(18)62021-0 [DOI] [Google Scholar]
- Wang M., Liu B., Li Y., Luo X., Li L. (2019). Non–target site based resistance to the ALS-inhibiting herbicide mesosulfuron-methyl in American sloughgrass (Beckmannia syzigachne). Weed Sci. 67 (5), 527–533. 10.1017/wsc.2019.22 [DOI] [Google Scholar]
- Yu Q., Powles S. (2014. a). Metabolism-based herbicide resistance and cross-resistance in crop weeds: a threat to herbicide sustainability and global crop production. Plant Physiol. 166 (3), 1106–1118. 10.1104/pp.114.242750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q., Powles S. B. (2014. b). Resistance to AHAS inhibitor herbicides: current understanding. Pest Manag. Sci. 70 (9), 1340–1350. 10.1002/ps.3710 [DOI] [PubMed] [Google Scholar]
- Yuan J. S., Tranel P. J., Stewart C. N., Jr. (2007). Non-target-site herbicide resistance: a family business. Trends Plant Sci. 12 (1), 6–13. 10.1016/j.tplants.2006.11.001 [DOI] [PubMed] [Google Scholar]
- Zheng D., Kruger G. R., Singh S., Davis V. M., Tranel P. J., Weller S. C., et al. (2011). Cross-resistance of horseweed (Conyza canadensis) populations with three different ALS mutations. Pest Manag. Sci. 67 (12), 1486–1492. 10.1002/ps.2190 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw Illumina sequence reads have been deposited in the NCBI Sequence Read Archive (SRA) database. The accession numbers for the samples with herbicide treatment were SRR10737896 and SRR10737895. And the accession numbers for the samples without herbicide treatment were SRR10742660 and SRR10742659.