Figure 1.
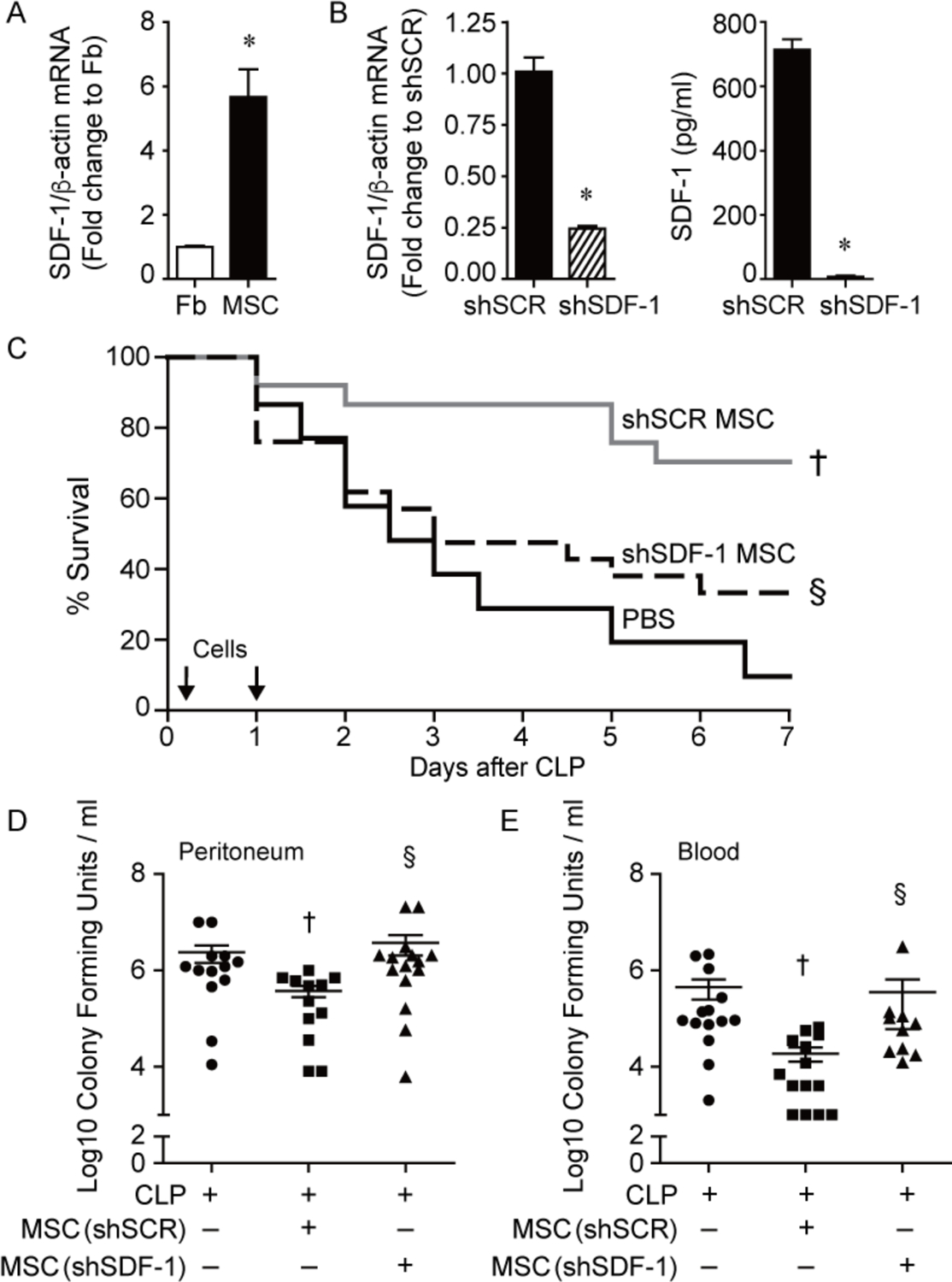
Improved survival and bacterial clearance by MSCs during polymicrobial sepsis is lost after silencing SDF-1. A) RNA was extracted from mouse lung fibroblasts (Fb, white bar) and bone-derived MSCs (black bar) and quantitative real-time PCR (qRT-PCR) was performed for SDF-1. Data are presented as fold change in RNA levels of SDF-1 normalized to β-actin, mean ± SEM, n=6 per group from three independent experiments. * versus Fb, P=0.0003. B) RNA was extracted from shSCR MSCs (black bar) and shSDF-1 MSCs (striped bar), and qRT-PCR was performed for levels of SDF-1. Data are presented as fold change in RNA levels of SDF-1 normalized to β-actin, mean ± SEM, n=9 per group from three independent experiments. * versus shSCR, P<0.0001. ELISAs for SDF-1 were also performed on cell culture supernatants of shSCR and shSDF-1 MSCs. * versus shSCR, P<0.0001. C) Septic BALB/c mice were randomly separated into 3 groups: PBS control (black solid line, n=15), shSCR MSCs (gray solid line, n=25), and shSDF-1 MSCs (black dashed line, n=25). All mice were subjected to CLP, and 2 hours after CLP the mice received PBS or cells (5 × 105) via tail vein injection. This treatment was also repeated at 24 hours (2.5 × 105) after CLP. Survival of mice was monitored over 7 days, and data are presented as Kaplan-Meier survival curves. † shSCR MSCs versus PBS, P=0.0006; § shSDF-1 versus shSCR, P=0.01. Bacteria were assessed in the peritoneum (D) and blood (E) in the PBS control (circles, n=13 or 14 respectively), shSCR MSCs (squares, n=12 or 14 respectively), and shSDF-1 MSCs (triangles, n=15 or 11 respectively). P=0.0077 and P=0.0005 respectively, with significant comparisons † versus CLP+PBS (− MSCs), § versus CLP+shSCR MSCs.
