We and others recently showed that coordinated expression of long non-coding (lnc)RNA is essential for myeloid differentiation and that, in turn, deregulation of lncRNA contributes to the pathogenesis of acute myeloid leukemia (AML).1 To better understand the relationship between lncRNA that control granulopoiesis and the block of differentiation in AML blasts, we sequenced stranded, non-poly-A-enriched cDNA libraries that were prepared from healthy human donor bone marrow (deposited under GEO accession number #GSE98946). Within this dataset, expression of the lncRNA Cancer Susceptibility 15 (CASC15) was significantly inversely correlated with myeloid differentiation and highly enriched in myeloblasts,1 which suggests a role in maintaining stem cell features such as an immature phenotype and/or self-renewal. Due to the reported proto-oncogenic role of CASC15 in childhood AML,2 we further explored CASC15 in the context of adult AML, especially in patients with mutated isocitrate dehydrogenase (IDH).
Figure 1.
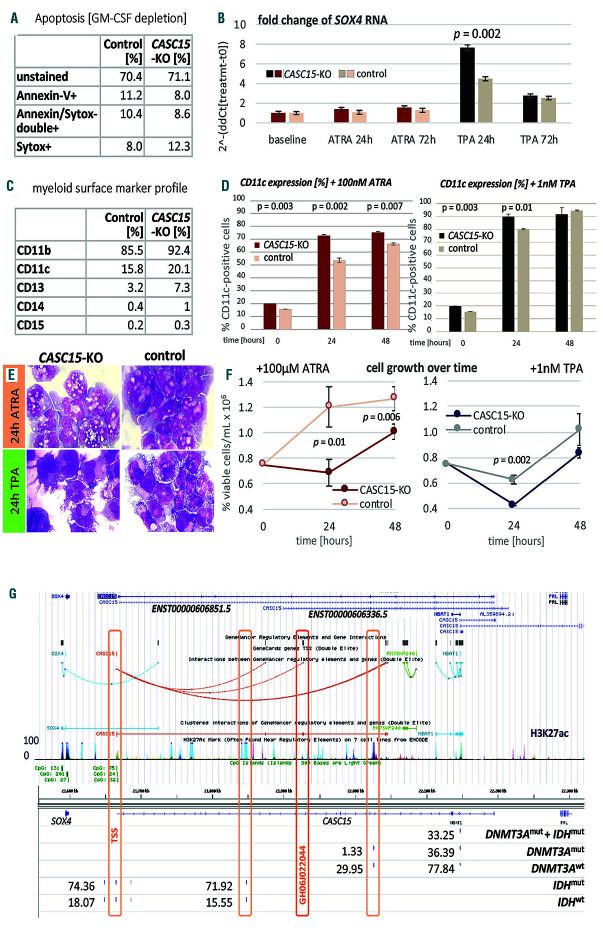
CASC15-KO promotes the differentiation of acute myeloid leukemia cells. (A) Apoptosis in CASC15-KO and empty vector-transduced (control) OCI-AML5 cell lines after 24 h of depletion of granulocyte-macrophage colony-stimulating factor (annexin-FITC/Sytox blue flow cytometry). (B) Expression of SOX4 during in vitro differentiation of CASC15-KO and control OCI-AML5 cell lines. All cells were treated with 0.1 mM all-trans retinoic acid (ATRA) and 1 nM 12-O-tetradecanoylphorbol-13-acetate (TPA) over 72 h in three independent experiments. Total RNA was extracted before, after 24 h and after 72 h of treatment, DNase-digested and transcribed to cDNA. A quantitative real-time polymerase chain reaction (qRT-PCR) was performed using SYBR green chemistry with subsequent melting curve analysis in technical triplicates. The 2-ddCt was calculated relative to the pre-determined housekeeping gene encoding succinate dehydrogenase complex subunit C (SDHC). (C) Baseline expression of the monocyte/macrophage markers CD11b (integrin subunit alpha M, ITGAM), CD11c (integrin subunit alpha X, ITGAX), and CD14, the granulocyte marker CD15 (fucosyltransferase 4, FUT4), and the general myeloid marker CD13 (aminopeptidase N, APN) in CASC15-KO and control cells. The percentages of positive cells, quantified by flow cytometry after 72 h, are shown. (D-F) Growth rate and CD11c myeloid cell surface marker expression of CASC15 and control cell lines during drug-induced in vitro differentiation (D) Cells (0.75x106/mL) were seeded and treated with 0.1 mM ATRA, 1 nM TPA or 0.1 mM vehicle control (dimethylsulfoxide) for 48 h in three independent experiments. CD11c was stained and quantified before (baseline/t0), after 24 h and after 48 h of treatment on 10,000 cells per sample by flow cytometry. (E) Cellular morphology was assessed in cytospins stained with May-Grünwald/Giemsa. (F) Number of trypan blue-negative cells 24 h and 48 h after drug treatment. (Figure continued on the next page) (G) The SOX4, CASC15, and NBAT1 genomic loci displayed in the University of California, Sant Cruz (UCSC) genome browser with subordinated tracks showing currently annotated isoforms (GENCODE v29), GeneHancer-listed regulatory elements, layered H3K27 acetylation (ENCODE), 5‘-C-phosphate-G-3‘ (CpG) sites, and percent methylation determined via enhanced reduced representation bisulfite sequencing (ERRBS) in IDH-mutant (n=9), DNMT3A-mutant (n=16), and IDH/DNMT3A double-mutant (n=11) AML patients relative to normal controls. The ERRBS data were produced and normalized by Glass et al.8 (Gene Expression Omnibus identiy: GSE86952). P-values were calculated using a paired Welch two sample t-test after testing for unequal variance.
To assess the role of CASC15 in human AML cells, we genetically deleted the promoter and transcription start site of CASC15 in the human OCI-AML-5 cell line via a CRISPR/Cas9 approach (CASC15-KO). In brief, OCIAML- 5 cells were transduced with Cas9-ORF-carrying a DHC013 vector and selected with 5 mg/mL blasticidin. Cas9 expression was confirmed by western blotting. Single guide (sg)RNA were designed using CCTop (https://crispr.cos.uni-heidelberg.de/): T1: 5’-CACCGATGGATTACAATTTGATCGC- 3’fw, 5’-AAACGCGATCAAATTGTAATCCATC- 3’rev; T2: 5’-CACCgATTAATTAAAGTTAGCTTCC-3’fw, 5’- AAACGGAAGCTAACTTTAATTAATC-3’rev. Each sgRNA pair was cloned into either pL40CCRISPR. EFS.PAC or pL-CRISPR.EFS.tRFP, respectively (kindly provided by Dirk Heckl), each with the Cas9-ORF removed. Cells (1x106/mL) were spin-infected with an equimolar mix of virus-containing medium + 10 mg/mL polybrene. Single-cell clones were sorted, expanded in 1 mg/mL puromycin, and sent for Sanger sequencing. Lack of CASC15 expression was confirmed in heterozygous knock-out (KO) and control cell lines via polymerase chain reaction (PCR) (PlatinumTaq, ThermoFisher Scientific) and quantitative real-time PCR (qRT-PCR) (TBgreen, TaKaRa, Japan) using the following primer sets: CASC15 exon1: 5’-GGGTATCTCCCTCTCGCAAC- 3’fw, 5’-CGTTCGGACACTTTTTCCCG-3’rev; CASC15 exon2: 5’-TGACCTCCTTCATTCTGCGTT- 3’fw, 5’-GGGTATAAGCCCCAGACCAA-3’rev. Loss of CASC15 did not sustainably affect apoptosis or SOX4 RNA levels in these cells (Figure 1A, B), as was previously reported for CASC15-KO in a RUNX1-rearranged AML background,2 but increased the expression of myeloid surface markers (Figure 1C). To induce differentiation, we treated 0.75x106 cells with 1 nM 12-O-tetradecanoylphorbol- 13-acetate (TPA) or 100 nM all-trans retinoic acid (ATRA) (Sigma-Aldrich) and assessed the level of differentiation by flow cytometry using α-CD11b-APC (#17-0118-41, clone ICRF44, eBioscience, San Diego, CA, USA), α-CD11c-APC (#17-0116-42, clone 3.9, ThermoFisher Scientific, USA), α-CD13-FITC (#ab52461, clone WM15, abcam, Cambridge, UK), and α-CD14-APC (#17-0149-42, clone 61D3, eBioscience), α-CD15-FITC (#555401, lot 13878, Becton Dickinson, San Jose, CA, USA) antibodies.
In response to TPA and ATRA, CASC15-KO cells showed enhanced myeloid differentiation and reduced proliferation compared to the empty vector control cells (Figure 1D-F), suggesting that the presence of CASC15 contributes to the block of differentiation in human AML cells.
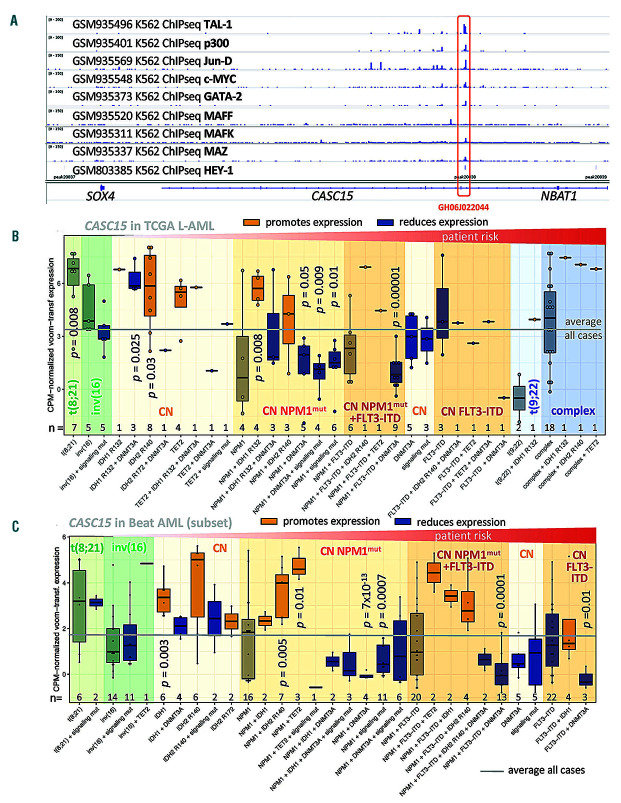
The highly complex genomic structure of the CASC15 locus pointed towards an equally complex transcriptional regulation and post-transcriptional processing of this (lnc)RNA (Figure 1G). Interrogating the entire transcription factor and co-factor footprinting data generated for the ENCODE project in K562 cells,4 we observed that the strongest binding intensities at the CASC15 site were for the erythroid differentiation factor TAL-1, the protooncogenic proteins JunD and c-MYC/MAZ, the co-activator p300, as well as for HEY-1, GATA-2, and members of the MAFF/BZIP family (Figure 2A). These transcription factors predominantly occupy the annotated CASC15 enhancer GH06J022044, which is located in intron 7 (GeneHancer v2, Jan 2019) (Figure 1G), share a strong enrichment for the basic helix-loop-helix motif, and are associated with transcriptional control in non-committed myeloid progenitors.5 This suggests that CASC15 is regulated by myeloid transcription factors, supporting our findings in human myeloblasts and AML cells.
Figure 2.
CASC15 is bound by hematopoiesis-regulating transcription factors and its expression reflects the patients’ IDH mutational status. (A) Chromatin immunoprecipitation sequencing (ChIPseq) data showing the location and intensity of transcription factor binding at the CASC15 genomic site, mapped to the hg19 human genome release. Data were produced and processed within the ENCODE project6 (Gene Expression Omnibus accession identities: GSE31477 and GSE32465). (B) Expression of CASC15 in a subset of the TCGA L-AML cohort12 (n=122), excluding MLL-rearranged, t(15;17), and aneuploid cases, subdivided according to recurrent cytogenetic and molecular abnormalities. RNA-sequencing data were TMM-normalized and log2-counts per million-transformed (limma).13 P-values were calculated comparing the individual groups to all cases using the Welch two-sample t-test and adjusted for multiple hypothesis testing. (C) Expression of CASC15 in a subset of t(8;21), inv(16), and cytogenetically normal patients with acute myeloid leukemia (AML) from the Beat AML Master Trial20 subdivided according to the recurrent cytogenetic and molecular abnormalities of interest. RNA sequencing data were TMM-normalized and log2- counts per million-transformed (limma).13 P-values were calculated comparing the individual groups to all cases using the Welch two-sample t-test and adjusted for multiple hypothesis testing.
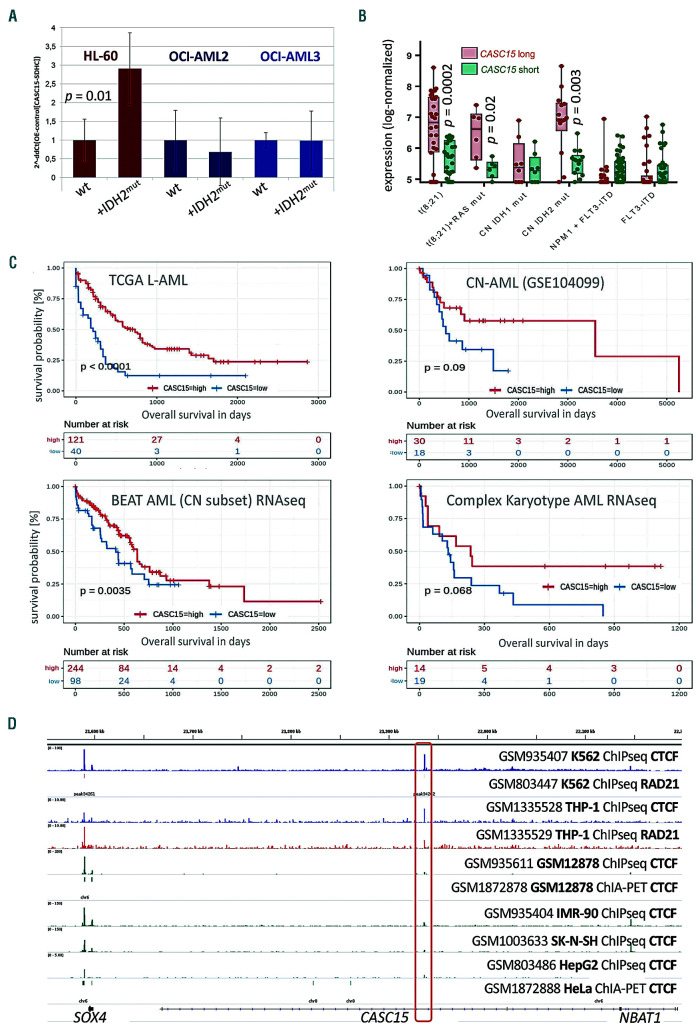
In four independent transcriptome datasets (including the Beat AML and TCGA LAML cohorts as well as GEO datasets #GSE6891 and #GSE104099, for a total of 1,145 patients), we observed the highest CASC15 expression in AML patients with t(8;21) and in cytogenetically normal AML patients harboring IDH or TET2 mutations (shown for the 2 largest cohorts in Figure 2B, C). Mutations in IDH1/2 and TET2 represent early, almost mutually exclusive events in AML patients and are associated with a hypermethylation genotype as both mutations eventually reduce the cytosine demethylation activity of TET2.6 In contrast, most DNMT3A mutations are thought to result in genome-wide CpG hypomethylation by formation of non-functional mutant:wild-type DNMT3A heterodimeric complexes.7 IDH and DNMT3A mutations frequently co-occur in AML and genomes of IDH/DNMT3A double-mutant patients were found to exhibit an epigenetic antagonism, in which CpG that are affected by either mutation alone are no longer affected.8 Interestingly, we observed strong downregulation of CASC15 in patients carrying mutant DNMT3A or signaling- activating mutations (FLT3, NRAS, KRAS, PTPN11) (Figure 2B, C). We confirmed this by engineered overexpression of IDH2R172K mutant cDNA in human DNMT3Awild-type (HL-60) and DNMT3Amutant (OCI-AML2/3) AML cell lines. The IDH2R172K-mutation was introduced into human IDH2wild-type cDNA via PCR (pEX-C0462-M02-50, Genecopoeia), cloned into a third-generation “selfinactivating” vector (kindly provided by Tobias Mätzig), and confirmed by Sanger sequencing. Lentiviral production and transduction were performed as previously described.3 Whereas CASC15 levels did not change in DNMTAR635W OCI-AML2 and DNMTAR882C OCI-AML3 cells, CASC15 was significantly upregulated (P=0.01) in DNMT3Awild-type HL-60 cells in response to ectopic expression of mutant IDH2 (Figure 3A).
Figure 3.
Expression of CASC15 is associated with patients’ survival, is induced by IDH mutations and antagonized by mutant DNMT3A. (A) Expression of CASC15 in IDH2R172K cDNA-overexpressing DNMT3A wild-type (HL-60) and DNMT3A-mutant (OCI-AML2 and OCI-AML3) cell lines. Lentivirus-production and infection were performed in two independent experiments each and confirmed by flow cytometry. Total RNA was extracted, DNase-digested and transcribed to cDNA. A real-time quantitative polymerase chain reaction was performed using SYBR green chemistry with subsequent melting curve analysis in technical triplicates. The 2-ddCt was calculated relative to the pre-determined housekeeping gene SDHC. (B) Expression of two CASC15 transcript variants in bone marrow from patients with acute myeloid leukemia (AML), subdivided according to recurrent cytogenetic and molecular abnormalities of interest. CASC15 long (red) shows log-normalized microarray data for probe 241047_at, which detects a short CASC15 transcript isoform (ENST00000606336.5). CASC15 short (green) shows log-normalized microarray data for probe 229280_s_at, detecting the longest CASC15 transcript isoform including all currently annotated exons (ENST00000606851.5). Data were produced and normalized by Verhaak et al.14 [Gene Expression Omnibus (GEO) accession identity: GSE6891]. P-values were calculated using a paired Welch two-sample t-test with 95% confidence and adjusted for multiple hypothesis testing. (C) Graphical representation of Kaplan-Meier estimates for the anti-differentiation transcript CASC15 calculated in three publicly available and one in-house generated (CN-AML) cohort of adult AML patients. The patients in each cohort were dichotomized into those with CASC15 high (red) and low (blue) expression using maximally selected rank statistics.15 Right-censored Kaplan-Meier estimates for overall survival were calculated using the R survival package with standard parameters. P-values were calculated using the log-rank test. (Figure continued on the next page) (D) AML cells possess a potential CTCF:RAD21-marked site that is not present in lymphoid and epithelial cells. CTCF-mapped chromatin interaction analysis by paired-end tag sequencing data were generated in GM12878 and HeLa cells by Tang et al. (GEO accession identity: GSE72816); chromatin immunoprecipitation sequencing (ChIPseq) data for CTCF and Rad21 in the THP-1 AML cell line were generated and analyzed by Rousseau et al. (GEO accession identity: GSE55407); ChIPseq data for CTCF and Rad21 in K562 cells were generated within the ENCODE project (GEO accession identity: GSE55407).4
A more detailed analysis of CASC15 expression in subgroups of AML patients with different cytogenetic and molecular features revealed a generally balanced expression of two different CASC15 isoforms: a short 7-exon (ENST00000606336.5) and a long CASC15 transcript containing all 12 annotated exons (ENST00000606851.5) (Figure 1G). However, in IDH2mutant as well as in t(8;21) patients, in whom CASC15 expression is highest, we detected a significantly (P<0.003) enriched expression of its long isoform (Figure 3B). This isoform switch was not observed in groups of patients with low CASC15 expression, such as cytogenetically normal AML with FLT3-ITD (Figure 2B). This novel observation suggests differential or aberrant splicing in these subgroups of patients, with as of yet unknown cause or functional relevance.
In line with the elevated expression of CASC15 in lower-risk cytogenetic and molecular AML subtypes, we found that high CASC15 RNA expression was associated with improved overall survival in four independent adult AML datasets (n=584 patients) (Figure 3C).
Although we found CpG methylation to be generally increased in IDHmut and decreased in DNMT3Amut AML patients at the CASC15 transcription start site and gene body (Figure 1G), the finding that CASC15 is highly expressed in hypermethylated (IDHmut and TET2mut) and less expressed in patients with hypomethylated genotypes (DNMT3Amut and NPM1mut) suggests that these methylation changes do not directly affect CASC15 transcription. The molecular consequences of IDH and DNMT3A mutations are not limited to epigenetic changes but also affect cellular metabolism and disturb DNA damage responses,9,10 which is thought to promote genomic instability and to account for the high number of co-mutations observed in IDH- and DNMT3A-mutant leukemias. In comparison to the high CASC15 levels in IDH2R140 AML patients, we found that CASC15 expression was often lower in IDH1R132 and IDH2R172 patients (Figure 2B, C), which underlines the presence of different transcriptional landscapes in each mutational entity10 and suggests that the R140 and R172 mutations affect the cooperation of IDH2 with different interaction partners.
Finally, we also observed that the CASC15 genomic site possesses a potential topologically-associating domain boundary in THP-1 and K562 myeloid leukemia cells, which is located between two CASC15 enhancers and is not present in non-myeloid cell lines (Figure 3D). Aberrant methylation at this CTCF-insulator in response to methylation-affecting mutations could be a plausible mechanism responsible for the counterintuitive expression of CASC15 in hypo- versus hyper-methylated AML, as had already been shown in IDH-mutant gliomas,11 and could potentially affect regional splicing.
In summary, we report that the survival-associated lncRNA CASC15 acts as a proto-oncogene in AML cells, in which it affects proliferation and differentiation. We further found that its expression is indirectly linked to DNA methylation-affecting mutations and succumbs to splicing, potentially through a mechanism that involves an intragenic lineage-specific CTCF site. These novel insights warrant further investigation, especially in the context of treatment response to IDH inhibitors.
Acknowledgments
The authors would like to thank Professor Hans-Jörg Fehling (Ulm University) for providing the IDH2R172K over-expression construct.
References
- 1.Schwarzer A, Emmrich S, Schmidt F, et al. The non-coding RNA landscape of human hematopoiesis and leukemia. Nat Commun. 2017;8(1):218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fernando TR, Contreras JR, Zampini M, et al. The lncRNA CASC15 regulates SOX4 expression in RUNX1-rearranged acute leukemia. Mol Cancer. 2017;16(1):126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azoitei N, Diepold K, Brunner C, et al. HSP90 supports tumor growth and angiogenesis through PRKD2 protein stabilization. Cancer Res. 2014;74(23):7125-7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gertz J, Savic D, Varley KE, et al. Distinct properties of cell-type-specific and shared transcription factor binding sites. Mol Cell. 2013; 52(1):25-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valledor AF, Borràs FE, Cellell-Young M, Celada A. Transcription factors that regulate monocyte/macrophage differentiation. J Leukoc Biol. 1998;63(4):405-417. [DOI] [PubMed] [Google Scholar]
- 6.Figueroa ME, Abdel-Whab O, Lu C, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18(6):553-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Russler-Germain DA, Spencer DH, Young MA, et al. The R882H DNMT3A mutation associated with AML dominantly inhibits wildtype DNMT3A by blocking its ability to form active tetramers. Cancer Cell. 2014;25(4):442-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glass JL, Hassane D, Wouters BJ, et al. Epigenetic identity in AML depends on disruption of nonpromoter regulatory elements and is affected by antagonistic effects of mutations in epigenetic modifiers. Cancer Discov. 2017;7(8):868-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guryanova OA, Shank K, Spitzer B, et al. DNMT3A mutations promote anthracycline resistance in acute myeloid leukemia via impaired nucleosome remodeling. Nat Med. 2016;22(12):1488-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inoue S, Li WY, Tseng A, et al. Mutant IDH1 downregulates ATM and alters DNA repair and sensitivity to DNA damage independent of TET2. Cancer Cell. 2016;30(2):337-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flavahan WA, Drier Y, Liau BB, et al. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature. 2016;529(7584):110-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cancer Genome Atlas Research Network Ley TJ, Miller C, Ding L, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCarthy DJ, Chen Y, Smyth Differential expression analysis of multifactor RNA-seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40(10):4288-4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verhaak RG, Wouters BJ, Erpelinck CA, et al. Prediction of molecular subtypes in acute myeloid leukemia based on gene expression profiling. Haematologica. 2009;94(1):131-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hothorn T, Lausen B. On the exact distribution of maximally selected rank statistics. Comput Stat Data An. 2003;43(2):121-137. [Google Scholar]