Fig. 4. DNA binding of cGAS is required for fork stalling after damage and drug resistance.
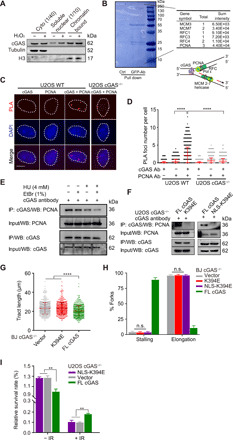
(A) Cytosolic, soluble nuclear, and chromatin fractions from U2OS cells were immunoblotted for cGAS and indicated proteins. (B) U2OS cells expressing GFP-cGAS were pulled down for interactome analysis. The results of mass spectrometry identified several major replication fork components. The schematic structure of cGAS pulled down with a replication fork complex via DNA or based on the proximity at replication forks. (C) Representative PLA of U2OS WT and cGAS−/− cells (scale bar, 10 μm). (D) Quantification of colocalization between cGAS and PCNA. Three independent experiments were done, mean ± SD is shown, and statistical analysis was done with the two-sided Mann-Whitney test; ****P < 0.001 and **** P < 0.0001. (E) Interaction of PCNA and cGAS detected with coimmunoprecipitation (Co-IP). U2OS cells were treated with or without 4 mM HU or 1% EtBr before Co-IP analysis with anti-cGAS antibody. (F) Interaction of cGAS mutants and PCNA detected with Co-IP. U2OS cGAS−/− cells were transfected with cGAS flag-tagged FL, K394E mutant, or NLS-K394E mutant for 36 hours before Co-IP analysis with anti-cGAS antibody. (G) Analysis of nascent DNA tract length (n = 600) in BJ cGAS−/− cells transfected with K394E mutant and corresponding control plasmids. For all experiments in this study, three independent experiments were done, mean ± SD is shown, and statistical analysis was done with the two-sided Mann-Whitney test; **** P < 0.0001. (H) DNA fiber analysis of replication fork stalling and elongation (n = 1800) in BJ cGAS−/− cells transfected with DNA binding defective mutants. Means and SD of three independent experiments are shown. (I) Colony formation assay for U2OS cGAS−/− cells expressing NLS-K394E mutants without and with 4-Gy IR treatment.
