Fig. 6. Rapid depletion of Pol I and diagram of two distinct suppression mechanisms of rRNA transcription.
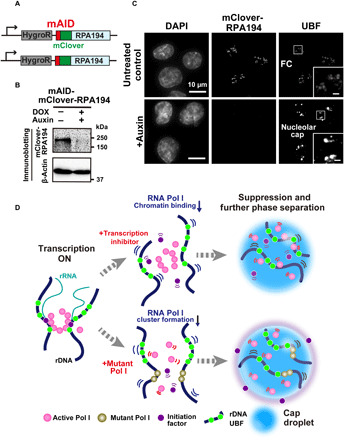
(A) Diagram of a genome-edited HCT116 line expressing mAID-mClover-RPA194 from the endogenous RPA194 gene locus. (B) Rapid Pol I degradation in HCT116 cells after auxin addition. (C) Localization of UBF in the cells before (first row, untreated control) and after rapid depletion of Pol I (second row, +auxin). Insets show enlarged images of the regions indicated with boxes. Scale bars, 1 μm. (D) Left: Active Pol I molecules form a stable cluster/condensate to transcribe rRNA genes (rDNA), constraining rDNA chromatin. Top: Once transcription is inhibited by a drug, the Pol I cluster/condensate detaches from chromatin, thus releasing the chromatin constraint. Pol I behaves like a liquid in the nucleolar cap through further phase separation. Bottom: Mutant Pol I stably binds to rDNA chromatin and inhibits WT Pol I cluster/condensate formation. The whole Pol I population becomes mobile in the nucleolar cap. An initiation factor is excluded from the cap, ensuring robust transcription suppression within the cap. Figure 6D consists of illustrations reproduced from figs. S3E, S4H, and S7A. Note that this model is highly simplified.
