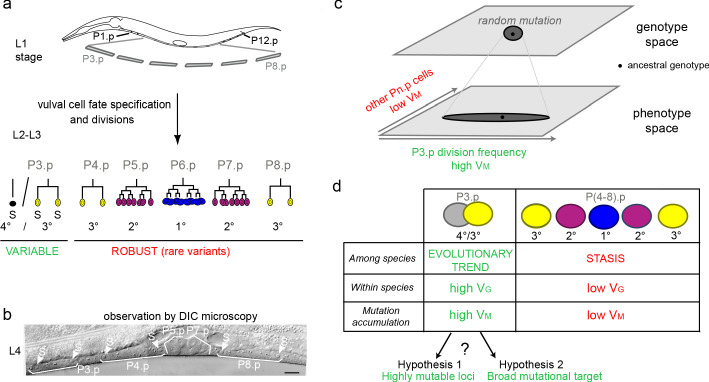
Figure 1. Specific evolutionary features of P3.p among vulva precursor cells and the question of the origin of its high mutational variance.
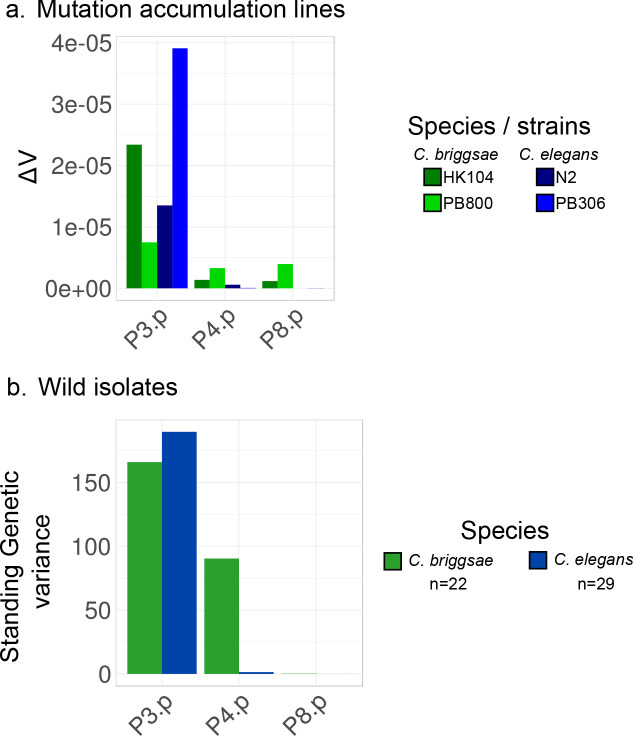
(a) Schematic description of development of the six vulva precusor cells (VPCs). The six cells P(3-8).p are born during the L1 larval stage. At the end of larval stage L2, P3.p either fuses with the surrounding hypodermal syncytium (hyp7) or escapes fusion like the other VPCs. The VPCs that have not fused divide in the L3 stage according to a fixed fate and lineage (1°, 2° and 3° fates, color-coded). (b) Nomarski picture of a mid-L4 stage animal showing the descendants of VPCs. In this individual, P3.p divides like P4.p and P8.p, as shown by the presence of two nuclei per mother Pn.p cell (labeled 'S' for syncytial). (c) Schematic genotype-phenotype map for the Pn.p cells, showing that P3.p has a high mutational variance. The black dot depicts the ancestral genotype and phenotype, and the dark grey shape schematizes the distribution after random mutation. (d) Unlike P(4-8).p, P3.p displays evolutionary change among Caenorhabditis species (evolutionary trend), a high polymorphism within species (standing genetic variance VG), and a high mutational variance (VM) found in mutation accumulation lines (Delattre and Félix, 2001; Braendle et al., 2010; Pénigault and Félix, 2011a). The high mutational variance of P3.p may be explained by a high mutation rate at specific loci or by a broad mutational target.