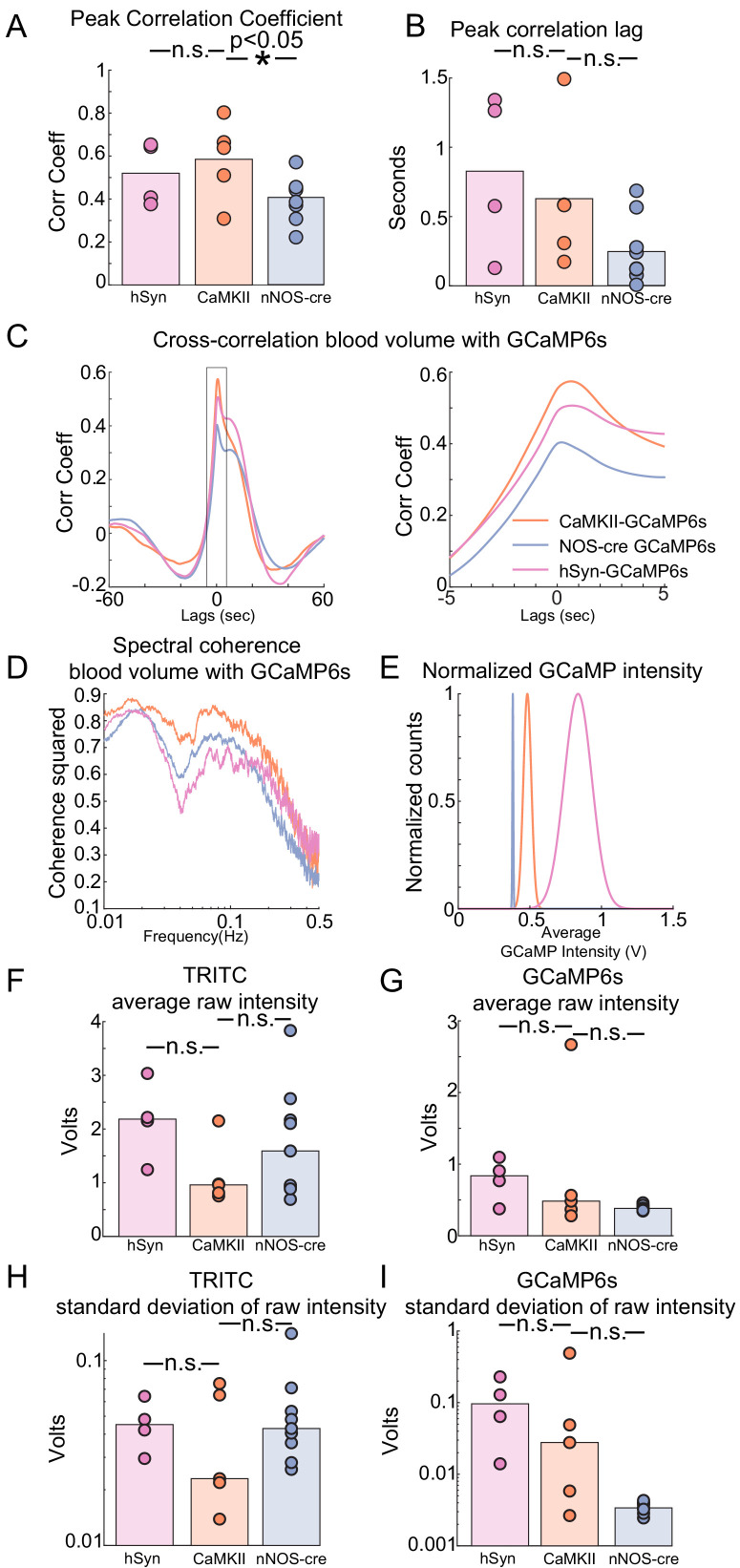
(A) Population averaged peak correlation coefficient between blood volume and GCaMP6s fluorescence signals across all behaviors. Bars are population mean of each GCaMP6s sub-type; circles are individual animal averages. hSyn: 0.54 ± 0.14, LME difference from CaMKII: p<0.7, CaMKII: 0.60 ± 0.17, nNOS: 0.43 ± 0.11, LME difference from CaMKII: p<0.02. (B) Population average, lag time of peak correlation coefficient between blood volume and GCaMP6s fluorescence signals across all behaviors. Bars are population mean of each GCaMP6s sub-type; circles are individual animal averages. hSyn: 0.79 ± 0.57 s, LME difference from CaMKII: p<0.60 CaMKII: 0.60 ± 0.52 s, nNOS: 0.24 ± 0.23 s, LME difference from CaMKII: p<0.09. (C) Population average traces of time lagged cross correlation between blood volume and GCaMP of ±60 s and ±5 s windows. (D) Population average traces of cross spectral frequency coherence between blood volume and GCaMP6s fluorescence signals. (E) Population average traces of normalized gaussian distribution of GCaMP6s fluorescence signals based on sub-type. (F) Population median of, average raw blood volume intensity across all behaviors. Bars are population median of each GCaMP6s sub-type; circles are individual animal averages. hSyn: 2.16 ± 0.73, LME difference from CaMKII: p<0.07 CaMKII: 1.13 ± 0.58, nNOS: 1.75 ± 1.03, LME difference from CaMKII: p<0.18. (G) Population median of, average raw GCaMP6s intensity across all behaviors. Bars are population median of each GCaMP6s sub-type; circles are individual animal averages. hSyn: 0.79 ± 0.30, LME difference from CaMKII: p<0.81 CaMKII: 0.87 ± 1.01, nNOS: 0.39 ± 0.04, LME difference from CaMKII: p<0.1. (H) Population median of, standard deviation of raw blood volume intensity across all behaviors. Bars are population median of each GCaMP6s sub-type; circles are individual animal averages. hSyn: 0.05 ± 0.01 s, LME difference from CaMKII: p<0.56, CaMKII: 0.04 ± 0.00.03, nNOS: 0.05 ± 0.04 s, LME difference from CaMKII: p<0.33. (I) Population median of, standard deviation of raw GCaMP intensity across all behaviors. Bars are population median of each GCaMP6s sub-type; circles are individual animal averages. hSyn: 0.11 ± 0.09, LME difference from CaMKII: p<0.68 CaMKII: 0.16 ± 0.21, nNOS: 0.003 ± 6×10−4, LME difference from CaMKII: p<0.08.