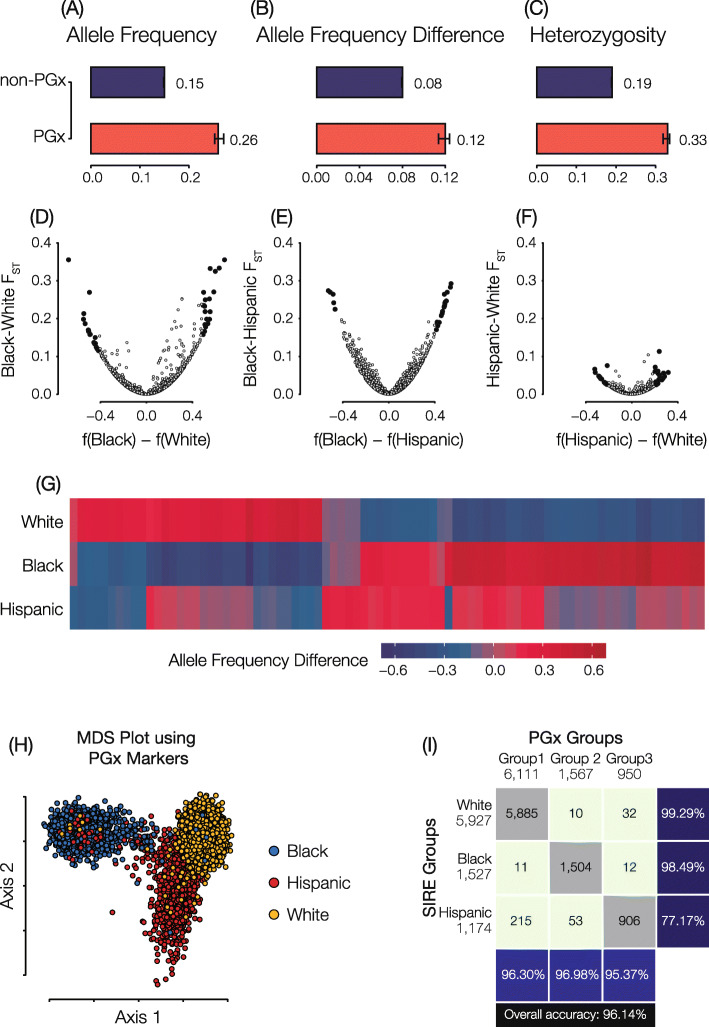
Fig. 2.
Pharmacogenomic variation in the US. Genome-wide average allele frequencies (a), group-specific allele frequency differences (b), and heterozygosity fractions (c) are shown for PGx variants (red) compared to non-PGx variants (blue). d–f Fixation index (FST; y-axis) and allele frequency differences (x-axis) for pairs of SIRE groups. Statistically significant PGx allele frequency differences are highlighted in black. g Heatmap showing group-specific allele frequencies for significantly diverged PGx variants. h Multi-dimensional scaling (MDS) plot showing the relationship among individual genomes as measured by PGx variants alone. Each dot is an individual HRS participant genome, and genomes are color-coded by participants SIRE. i The correspondence between SIRE groups and PGx groups defined by K-means clustering on the results of the MDS analysis. Data shown here correspond to SIRE groups; analogous results for GA groups are shown in Supplementary Figure 4 (see Additional file 1: Figure S4)