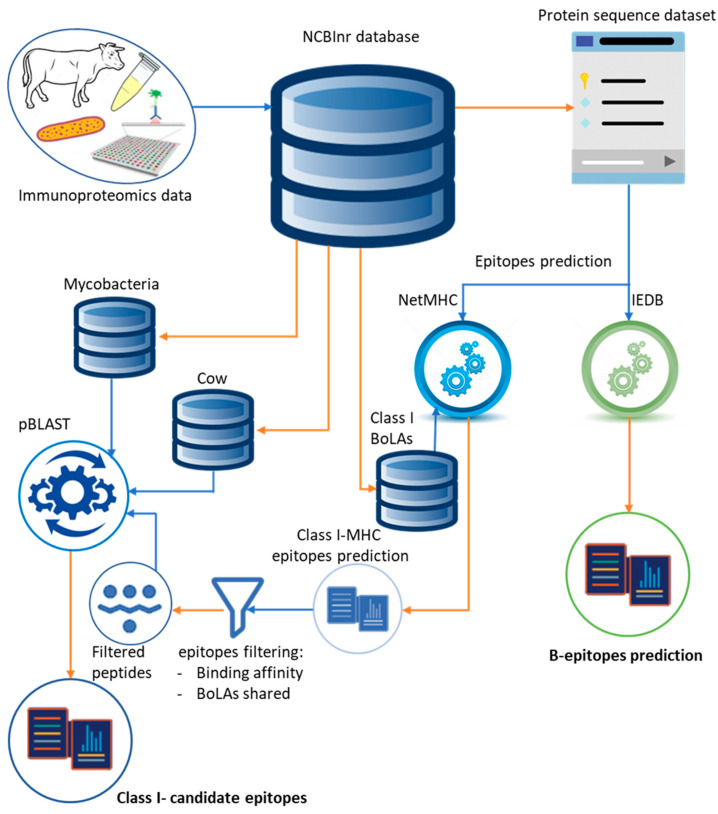
Figure 4.
Schematic representation of the immunoinformatic approach. Blue and orange arrows refer to the input and output of the data, respectively. Data arising from our previous immunoproteomic study [18] were used to retrieve the protein sequences of the immunogenic proteins, selected on the basis of their capability of being complexed by the immunoglobulins in the sera of the infected cows. The sequence of the immunogenic proteins is subjected to the epitope prediction through dedicated tools and algorithms. The B-epitope prediction was performed via the IEDB prediction tool, that provides a list of candidate B-epitopes. The class I MHC epitopes waspredicted via NetMHC. This makes use of the BoLA haplotypes from the data repository (NCBI) as references for computing the linear peptides capable of being recognized and presented by the diverse BoLAs. The list of potential T-epitopes is further refined by selecting the most commonly recognized epitopes with a relatively high binding affinity. The refined list of epitopes is further tested for sequence specificity and cross-reactivity by pBLAST alignment versus the mycobacteria and cow protein database which, in turn, arise from the publicly available data repository (NCBI).