Figure 1. Immune cells acquire can additional spacers upon infection.
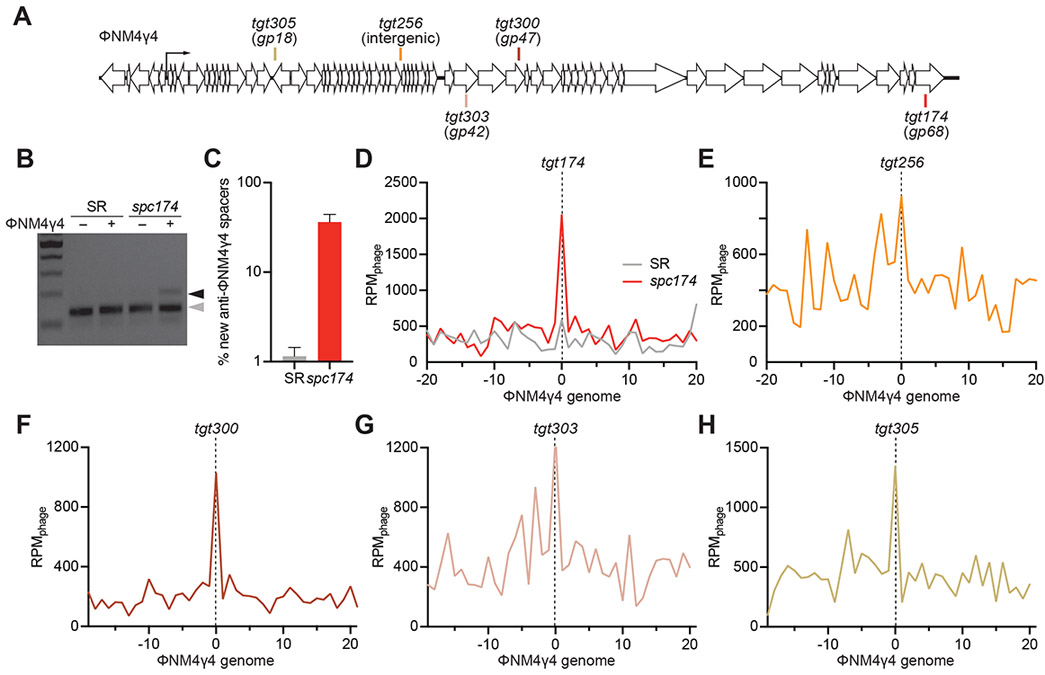
(A) Staphylococcal phage ΦNM4γ4 genome and the Cas9 targets analyzed in this study. (B) Staphylococci harboring pCRISPR(spc174) that targets the phage ΦNM4γ4 and pCRISPR(SR) were infected (+) or mock-infected (−) for 30 minutes and their DNA purified to amplify the CRISPR locus. PCR products were separated by gel electrophoresis to detect the acquisition of new spacers. Grey and black arrows: non-expanded and expanded, respectively, CRISPR arrays. (C) Fraction (%) of spacer sequences matching to the genome of phage ΦNM4γ4 detected in the PCR product of the expanded CRISPR array of staphylococci harboring either pCRISPR(spc174) or pCRISPR(SR). Mean ± StDev values of three independent experiments are shown. (D-H) Distribution of new spacer sequences detected 30 minutes after infection of staphylococci expressing Cas9 programmed to cleave the targets shown in (A). Reads per million of phage reads (RPMphage) are mapped to 1 kb bins of the ΦNM4γ4 genome (shown in linear form, with the specified target in the center). Average curve of three independent experiments is shown. See also Fig. S1.
