Figure 2. PAM, but not seed, target mutations, abrogate new spacer acquisition in immune cells.
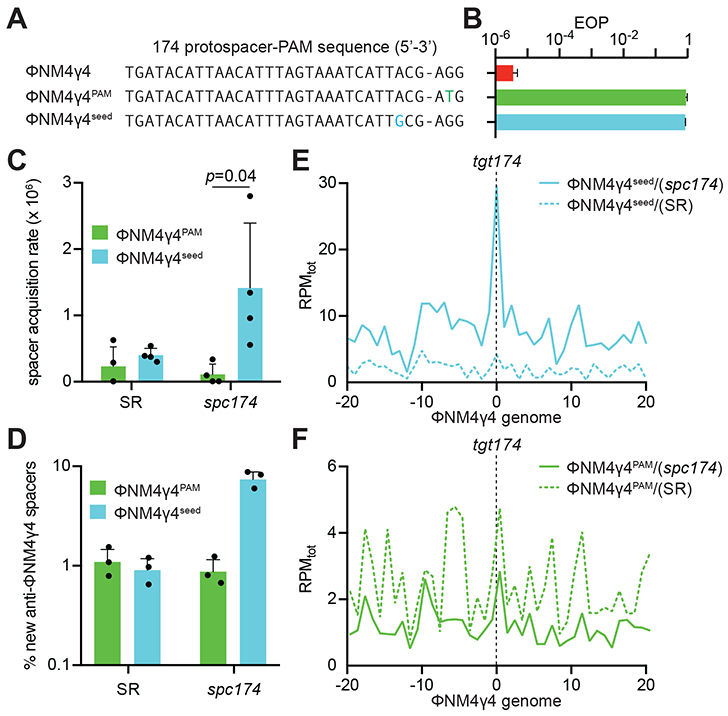
(A) ΦNM4γ4 variants containing different tgt174 sequences: wild-type, PAM (AGG>ATG) and seed (A−3>G−3). (B) Propagation ability of the three ΦNM4γ4 variants described in (A) on staphylococci harboring pCRISPR(spc174), measured as efficiency of plaquing (EOP). Mean ± StDev values of three independent experiments are shown. (C) Spacer acquisition rate after infection with different ΦNM4γ4 phages containing mutations in either the PAM or seed of tgt174, measured as the fraction of cells [harboring either pCRISPR(spc174) or pCRISPR(SR)] that survive infection through the acquisition of a new spacer. Mean ± StDev values of three independent experiments are shown. (D) Fraction (%) of spacer sequences matching to the genome of phage ΦNM4γ4 detected in the PCR product of the expanded CRISPR array of staphylococci harboring either pCRISPR(spc174) or pCRISPR(SR) after infection with different ΦNM4γ4 phages containing mutations in either the PAM or seed of tgt174. Mean ± StDev values of three independent experiments are shown. (E) Distribution of new spacer sequences detected in (D) in cells harboring pCRISPR(spc174), measured as spacer reads per million of total reads (RPMtot), and mapped to 1 kb bins of the ΦNM4γ4 genome (shown in linear form, with tgt174 in the center). Average curve of three independent experiments is shown. (F) Same as (E), but after analysis of bacteria harboring pCRISPR(SR). See also Figs. S2 and S3.
