Figure 3. Target orientation does not affect the pattern of spacer acquisition.
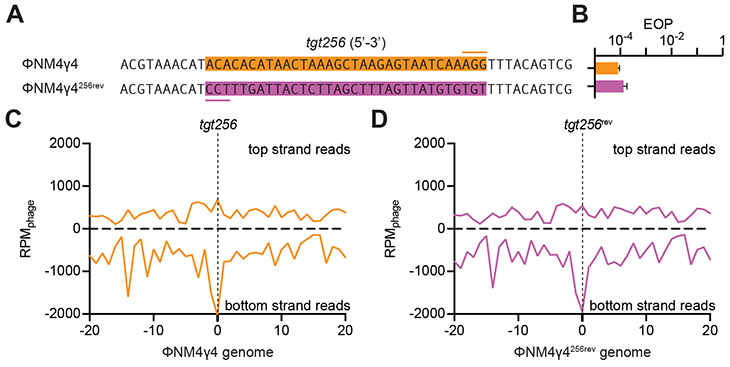
(A) The sequence of tgt256 on the ΦNM4γ4 genome (orange box, the line marks the PAM) was reversed in the ΦNM4γ4256rev mutant virus to allow annealing of the crRNA generated by spc256 to the other strand of the phage genome. (B) Comparison of the efficiency of plaquing (EOP) of ΦNM4γ4 on staphylococci carrying pCRISPR(spc256) of both phages shown in (A). Mean ± StDev values of three independent experiments are shown. (C) Distribution of new spacer sequences acquired after infection of staphylococci carrying pCRISPR(spc256) with ΦNM4γ4, measured as spacer reads per million of total phage reads (RPMphage), and mapped to 1 kb bins of either the top or bottom strands of the phage genome (shown in linear form, with tgt256 in the center). Average curve of three independent experiments is shown. (D) Same as (C) but after infection with ΦNM4γ4256rev. See also Fig. S4.
