Figure 4. Target DNA cleavage is required for type II primed spacer acquisition.
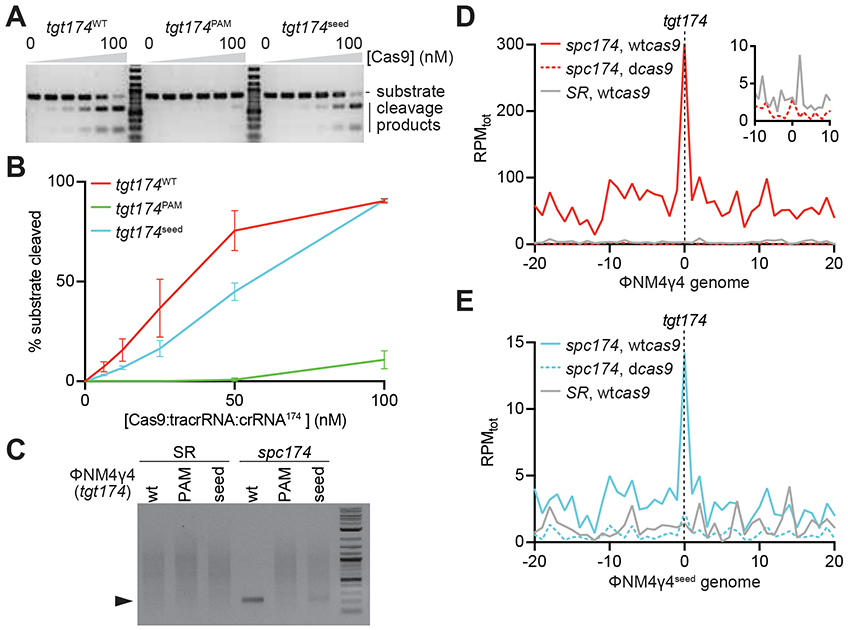
(A) In vitro cleavage assay of a dsDNA oligonucleotide containing the different tgt174 sequences shown in Fig. 2A, incubated with increasing concentrations of a 1:1:1 mix of Cas9:tracrRNA:crRNA174: 0, 6.25, 12.5, 25, 50 and 100 nM. Substrates and cleavage products were separated by agarose gel electrophoresis. A representative image of three replicates is shown. (B) The bands corresponding to the cleavage products in (A) were quantified and plotted against the concentration of the Cas9 ribonucleoprotein complex. Mean ± StDev values of three independent experiments are shown. (C) In vivo cleavage of phage DNA. Cells containing the pCRISPR(spc174) or pCRISPR(SR) plasmids where infected with ΦNM4γ4, ΦNM4γ4174-seed or ΦNM4γ4174-PAM phages and the cleavage products (marked by the black triangle) of the tgt174 sequence were amplified and separated by agarose gel electrophoresis. (D) Distribution of new spacer sequences acquired after infection of staphylococci carrying either pCRISPR(spc174, wtcas9), pCRISPR(spc174, dcas9) or pCRISPR(SR), with ΦNM4γ4, measured as spacer reads per million of total reads (RPMtot), and mapped to 1 kb bins of either the top or bottom strands of the phage genome (shown in linear form, with tgt174 in the center). Average curve of two independent experiments is shown. The inset shows the data from cells harboring pCRISPR(spc174, dcas9) or pCRISPR(SR) around the target site, with a different RPMtot scale. (E) Same as (D) but after infection with ΦNM4γ4seed. See also Fig. S4.
