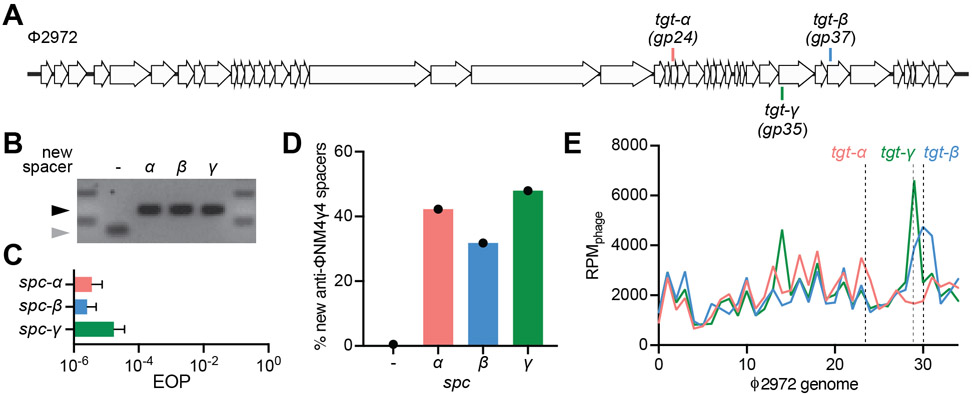
Figure 5. Cleavage-dependent spacer acquisition in Streptococcus thermophilus.
(A) Streptococcal phage Φ2972 genome and the Cas9 targets analyzed in this study. (B) Agarose gel electrophoresis of PCR products after amplification of the CRISPR3 array from S. thermophilus CRISPR3-naïve that acquired spacers α, β and γ upon infection with Φ2972. Grey and black arrows: non-expanded and expanded, respectively, CRISPR3 arrays. (C) Comparison of the efficiency of plaquing (EOP) of phage Φ2972 on CRISPR3-α, -β or -γ streptococci. Mean ± StDev values of three independent experiments are shown. (D) Fraction (%) of new spacer sequences matching to the genome of phage Φ2972 detected after infection of CRISPR3-α, -β or -γ streptococci. (−) indicates new spacers acquired in the absence of a targeting spacer by CRISPR-naïve cells. Values for a single experiment are shown. (E) Distribution of new spacer sequences detected 30 minutes after infection of streptococci expressing Cas9 programmed to cleave the targets shown in (A). Reads per million of phage reads (RPMphage) are mapped to 1 kb bins of the Φ2972 genome. Values for a single experiment are shown.