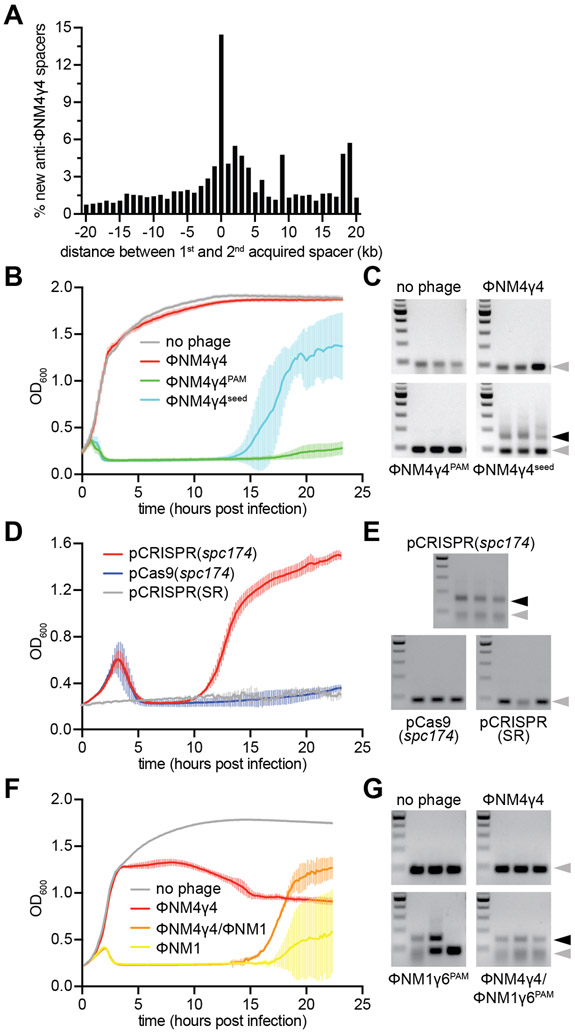
Figure 6. Cleavage-dependent spacer acquisition mediates a robust immune response.
(A) Distance between the targets in the ΦNM4γ4 genome specified by the first and second spacers acquired after infection of naïve staphylococci carrying the type II-A CRISPR-Cas system of S. pyogenes with ΦNM4γ4. The number of different spacers within 1-kb bins of the ΦNM4γ4 genome are shown; the position of first spacer acquired in each array is set as 0 kb. (B) Cell survival measured as OD600 after infection of cultures carrying pCRISPR(spc174) with different phages at MOI 10 or none as a control. The average curves of three different replicates are shown, with +/− StDev values shown in lighter colors. (C) Agarose gel electrophoresis of PCR products after amplification of the CRISPR array of cells obtained after the experiment in (B) to detect the integration of new spacers. Grey and black arrows: non-expanded and expanded, respectively, CRISPR arrays. (D) Cell survival measured as OD600 after infection with ΦNM4γ4 at MOI 100 of staphylococci carrying different plasmids. The average curves of three different replicates are shown, with +/− StDev values shown in lighter colors. (E) Same as (C) but for the cells obtained after the experiment in (D). (F) Cell survival measured as OD600 after infection of cultures carrying pCRISPR(spc174) with different phages at MOI 1 or none as a control. “ΦNM4γ4/ΦNM1” indicates infection with ΦNM1γ6PAM 30 minutes of addition of ΦNM4γ4 at MOI 10. “ΦNM1” is an abbreviation for ΦNM1γ6PAM. The average curves of three different replicates are shown, with +/− StDev values shown in lighter colors. (G) Same as (C) but for the cells obtained after the experiment in (F). See also Figs. S5 and S6.