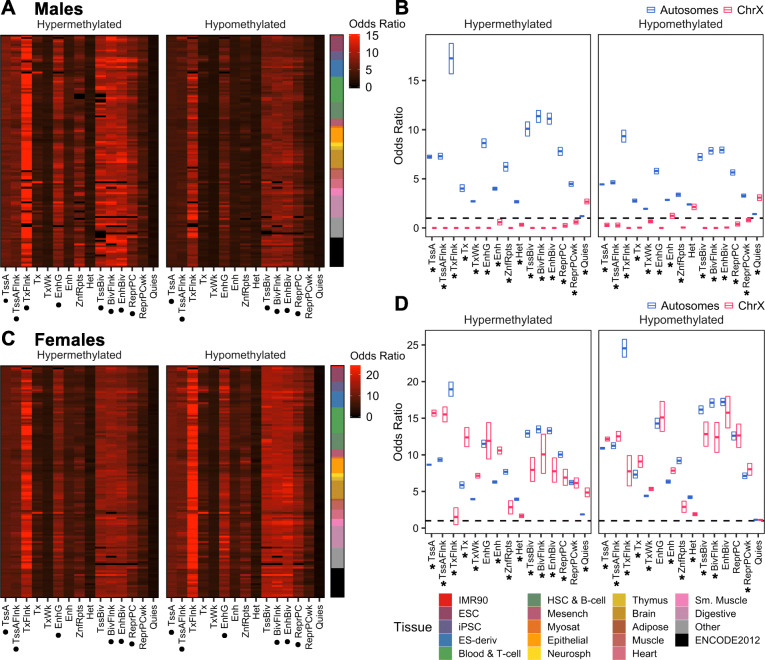
Fig. 5.
Pan-tissue chromatin signature of cord blood ASD DMRs reveals X-linked sex differences in chromatin states. Cord blood ASD DMRs from the discovery set were overlapped with 15-state model ChromHMM segmentations from 127 cell types in the Roadmap Epigenomics Project using the Locus Overlap Analysis (LOLA) R package. a, c The enrichment odds ratio was plotted for hypermethylated and hypomethylated DMRs identified in (a) males or (c) females from the discovery set. Top enriched (black dots) chromatin states were identified as those with odds ratio and −log(q-value) of at least the median value for that DMR set and with a q-value less than 0.05 for more than half of all cell types. b, d The enrichment odds ratio was plotted for hypermethylated and hypomethylated DMRs on autosomes or the X chromosome identified in b males or d females from the discovery set. Boxes represent mean and 95% confidence limits by nonparametric bootstrapping. Significance of differential enrichment of X chromosome compared to autosome DMRs was assessed by paired t-test of odds ratios for each cell type. P values were adjusted for the number of chromatin states using the FDR method (*q < 0.05, males TD n = 39, ASD n = 35; females TD n = 17, ASD n = 15). BivFlnk, flanking bivalent transcription start site or enhancer; Enh, enhancer; EnhBiv, bivalent enhancer; EnhG, genic enhancer; Het, heterochromatin; Quies, quiescent region; ReprPC, polycomb-repressed region; ReprPCWk, weak polycomb-repressed region; TssA, active transcription start site; TssAFlnk, flanking active transcription start site; TssBiv, bivalent transcription start site; Tx, strong transcription; TxFlnk, transcribed at gene 5′ and 3′; TxWk, weak transcription; ZnfRpts, zinc finger genes and repeats