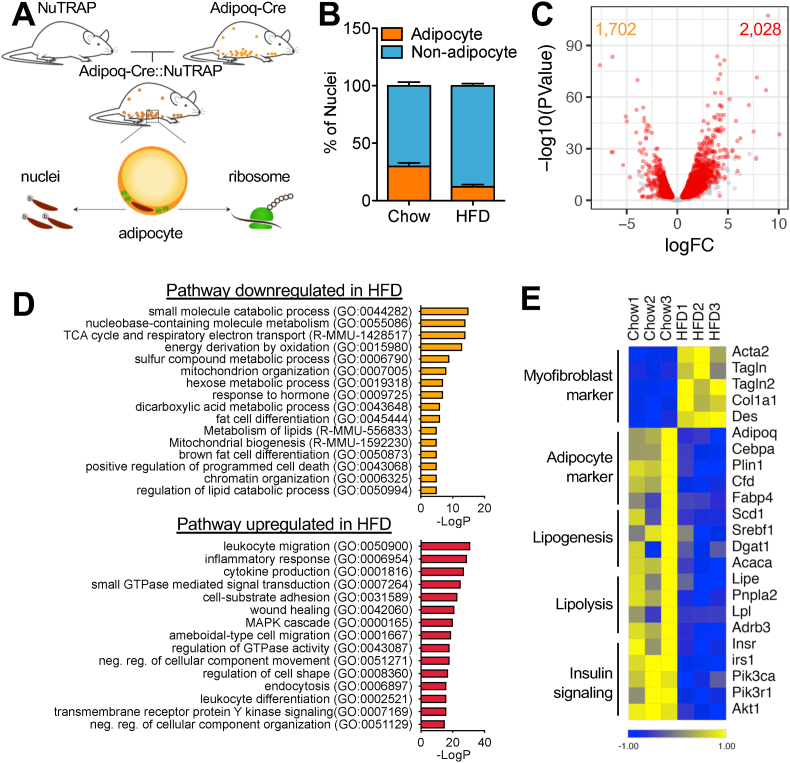
Figure 1.
NuTRAP/RNA-seq reveals aberrant alterations in molecular signature of adipocytes after HFD. (a) NuTRAP mice crossed with Adipoq-Cre mice to allow simultaneous labeling of adipocyte nuclei and ribosomes in adipocytes. (b) Adipocyte and non-adipocyte nuclei from eWAT of Adipoq-Cre::NuTRAP mice on chow and HFD were quantified by flow cytometry. Bars indicate mean ± SEM (n = 3–4 animals/group). (c) Volcano plot of the adipocyte-specific transcriptomes on chow and HFD by TRAP followed by RNA-seq (HFD/chow). Each dot indicates individual genes. Differentially regulated genes are shown in red (FDR ≤ 0.05, logFC ≥ 0.5, and logCPM ≥ 2). (d) GO pathway analysis with the differentially regulated genes. (e) Heatmap of myofibroblast and adipocyte markers and genes involved in adipocyte cellular metabolic processes. Columns represent biological replicates (n = 3 animals/group). Expression values (CPM) of each mRNA are represented by z scores.