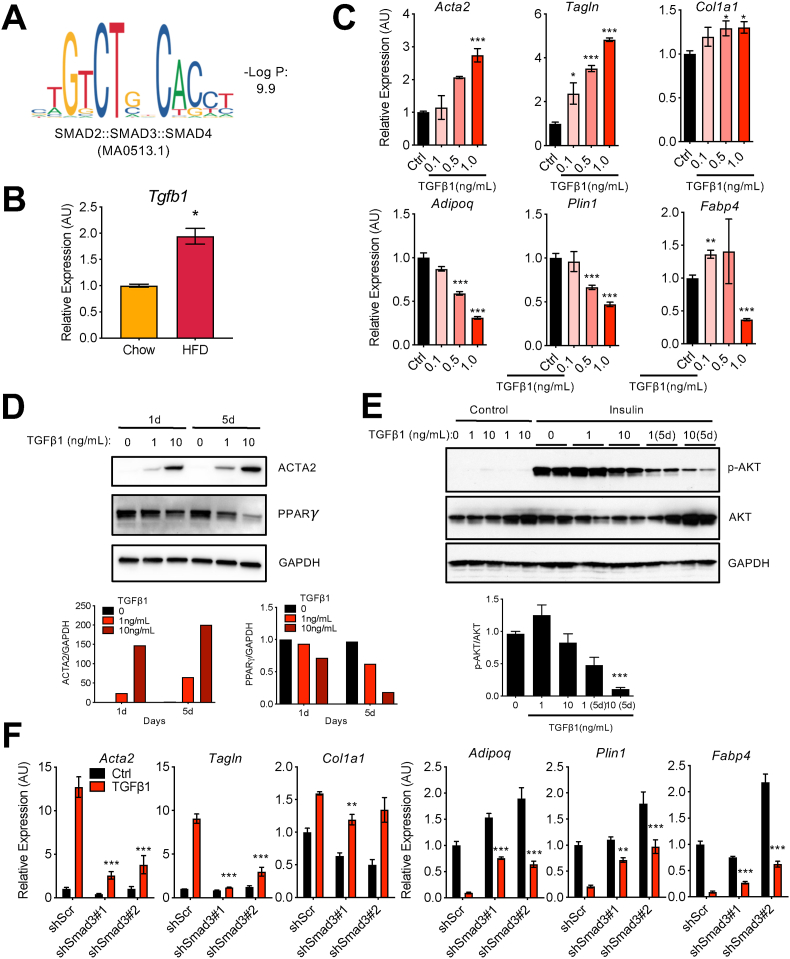
Figure 5.
TGFβ-SMAD3 pathway promotes aberrant changes in adipocytes in vitro. (a) SMAD2::SMAD3::SMAD4 motif over-represented in the H3K27ac peaks induced in adipocytes on HFD. (b) Tgfb1 mRNA expression in whole eWAT samples on chow and HFD by qRT-PCR analysis. Bars indicate mean ± SEM (n = 4–6 animals/group) (∗p < 0.05). (c) Gene expression analysis of the adipocyte and myofibroblast markers by qRT-PCR in differentiated 3T3-L1 adipocytes treated with TGFβ1 (1 ng/ml) for 24 h at the indicated concentrations. Bars indicate mean ± SEM (n = 3 replicates). (control vs TGFβ1 treatments; ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.005). (d) Protein levels of PPARγ and ACTA2 in differentiated 3T3-L1 adipocytes treated with TGFβ1 treatment for 24 h or 5 days at the indicated concentrations. GAPDH was used as a loading control. Bar graphs represent the quantification of ACTA2 and PPARγ protein levels relative to GAPDH. (e) Insulin signaling measured by analyzing AKT phosphorylation (p-AKT). Differentiated 3T3-L1 adipocytes were treated with TGFβ1 for 24 h or 5 days at the indicated concentrations, serum-starved for 4 h, and then stimulated with 100 nM insulin for 20 min. GAPDH was used as a loading control. Bar graph is the quantification of p-AKT/AKT by ImageJ (∗∗∗p < 0.005). (f) Effects of Smad3 knockdown on the expression of adipocyte and myofibroblast marker genes. Differentiated 3T3-L1 adipocytes were transduced with lentivirus expressing control scrambled shRNA or two different shSmad3 RNA for 4 days and treated with TGFβ1 (ng/mL) for 24 h. Bars indicate mean ± SEM (n = 3 replicates) (control with TGFβ1 vs shSmad3 with TGFβ1; ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.005).