-
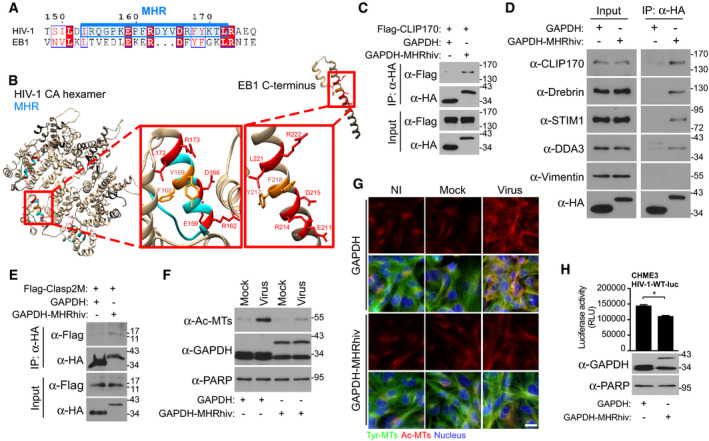
A
Sequence similarities of the EB1 C‐terminus with MHR (highlighted in blue) in HIV‐1 (NL4‐3). Identical residues are highlighted in red, and similar residues are printed in red.
-
B
Structure modeling of HIV‐1 capsid hexamers with focus on MHR alongside C‐terminus of EB1 in red boxes. Identical residues are indicated in red, and similar residues are indicated in dark orange. UCSF Chimera 1.12 software was used for structure modeling of NL4‐3 hexamer (PDB 3J3Q) and EB1 (PDB 2HKQ).
-
C–E
Anti‐HA co‐IP showing that GAPDH fused to the HIV‐1 MHR (GAPDH‐MHRhiv), but not HA‐tagged GAPDH (GAPDH) binds to Flag‐CLIP170 (C), endogenous forms of various human SxIP‐containing +TIPs, but not vimentin (D) or to Flag‐tagged SxIP fragment (Flag‐Clasp2M) (E) in 293T cells. Results are representative of three experimental replicates.
-
F, G
HIV-1‐VSV-luc (Virus), but not uninfected (NI) or Mock, infection induces the formation of Ac‐MTs in CHME3 expressing GAPDH‐MHRhiv, but not GAPDH, as shown by WB analysis (F) or imaging at 6 h.p.i (G). Scale bar, 15 μm. Representative fields are shown. Results are representative of three experimental replicates.
-
H
GAPDH‐MHRhiv expression in CHME3 reduces infection with HIV-1‐WT-luc. Statistical significance was determined by t‐test. *P < 0.05. Data are mean values from three independent experiments ± SEM.
Data information: Molecular weight markers (in kDa) are shown to the right of WBs.