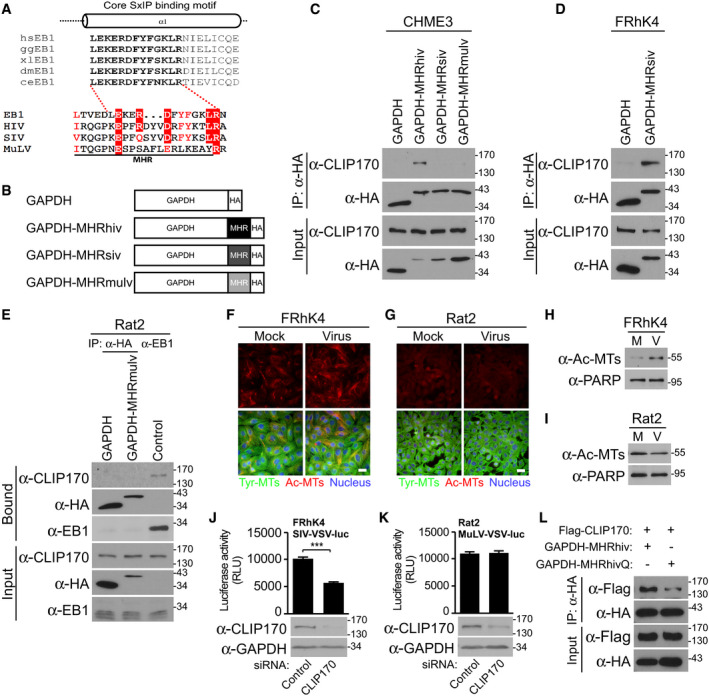
Figure 7. Differential EB1‐mimicry in MHR domains across retroviruses.
-
ASequence similarities of MHR in HIV‐1 (NL4‐3), SIVmac239, and MuLV with the core SxIP‐binding motif (bold text) within the EBH domain of EB1 from different species, adapted from Honnappa et al (2009). Residues identical to those in EB1 are highlighted in red, and similar residues are printed in red.
-
BSchematic representation of HA‐tagged GAPDH constructs, with MHR from HIV‐1 (GAPDH‐MHRhiv), SIVmac239 (GAPDH‐MHRsiv), or MuLV (GAPDH‐MHRmulv).
-
CAnti‐HA co‐IP in CHME3 expressing GAPDH‐MHRhiv, GAPDH‐MHRsiv, or GAPDH‐MHRmulv showing binding to endogenous CLIP170. Results are representative of three experimental replicates.
-
D, EAnti‐HA co‐IP in Rhesus macaque FRhK4 (D) or rat fibroblast Rat2 (E) cells expressing GAPDH alongside GAPDH‐MHRsiv or GAPDH‐MHRmulv, respectively. Anti‐EB1 co‐IP was included as positive control to demonstrate CLIP170 binding by EB1 in Rat2 cells. Results are representative of three experimental replicates.
-
F–IFRhK4 cells infected with SIV‐VSV-luc (F) or Rat2 cells infected with MuLV-VSV‐luc (G) stained for Ac‐MTs, Tyr‐MTs, and the nucleus (Hoechst) at 6 h.p.i, or analyzed by WB (H and I, respectively). Scale bar, 20 μm. Representative fields are shown. Results are representative of three experimental replicates.
-
J, KMeasurements of SIV‐VSV-luc or MuLV‐VSV-luc infectivity in FRhK4 (J) or Rat2 (K) cells depleted of CLIP170. Statistical significance was determined by t‐test. ***P < 0.001. Data are mean values from two independent experiments ± SEM.
-
L293T transfected with GAPDH‐MHRhiv or R162Q mutant (GAPDH‐MHRhivQ) was subjected to anti‐HA co‐IP. Representative WB analysis is shown (n = 2).
