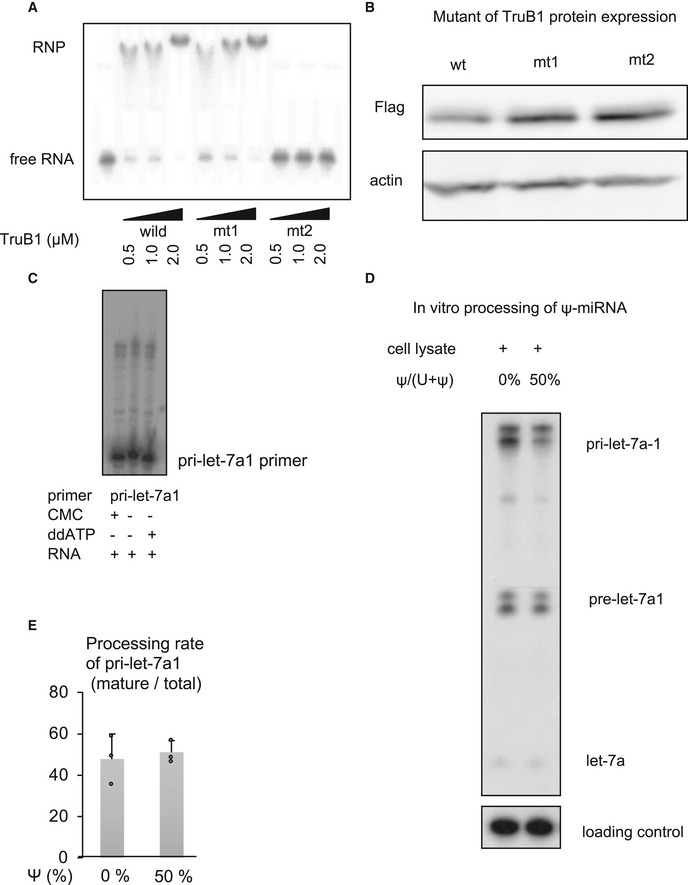
Figure EV2. Impact of pseudouridylation enzyme activity on microprocessing of let‐7.
-
AEMSA of 32p‐ATP-labeled tRNAPhe mixed with recombinant TruB1, mt1, or mt2 at several doses. RNP: Ribonucleoprotein complexes.
-
BProtein expression of Flag‐tagged TruB1 and its mutants. Protein expression was evaluated by Western blotting. Protein was isolated from HEK‐293FT cells infected with tetracycline‐inducible lentiviruses expressing TruB1, mt1, or, mt2, 5 days after doxycycline treatment.
-
CLocation of pseudouridine sites were detected by CMC primer extension. Total RNA purified from HEK‐293FT cells was treated with CMC. CMC‐treated RNA were reverse‐transcribed with RI‐labeled specific primers for pri‐let-7a1. ddATP was used as a sequence control.
-
D, EIn vitro processing analysis for pri‐let-7 with pseudouridine. 32p-ATP‐labeled pri‐let-7a1 was synthesized using UTP: pseudouridine at a ratio of 1:1 or 1:0. This labeled RNA was treated with whole cell lysate from TruB1 expressing HEK293FT cells transfected with pcDNA3.1‐TruB1. Autoradiographed image (D) and the relative processing rate (E) are shown. Error bars show SD; n = 3. Significance was assessed using 2‐tailed Student's t‐test.
Source data are available online for this figure.
