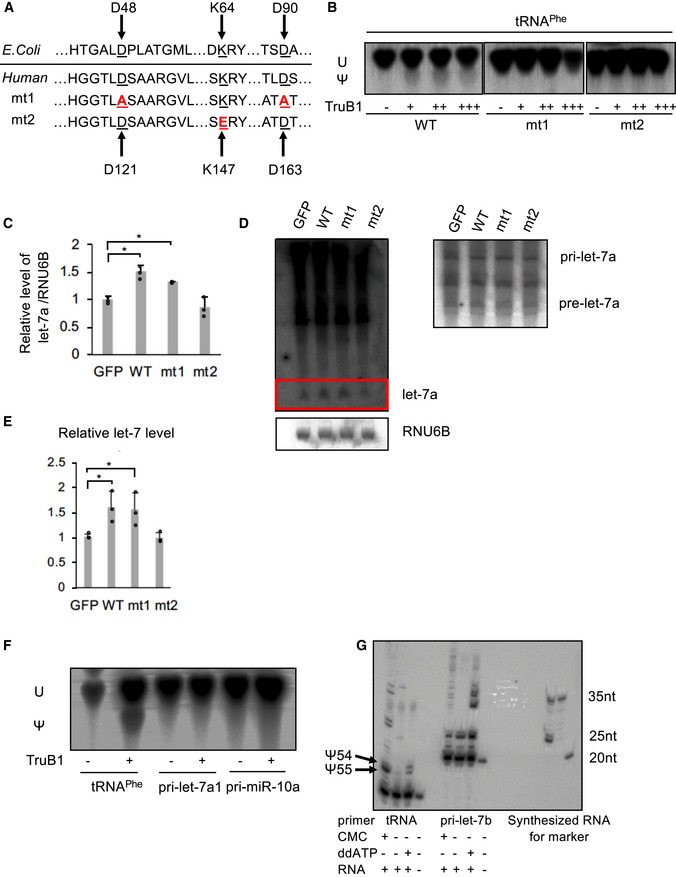
Amino acid sequences encoding for the enzyme activity and RNA‐binding ability in Escherichia coli TruB and human TruB1 (Top). Design of mutant 1 (mt1) and mutant 2 (mt2) (bottom).
In vitro enzyme activity assay. 32p‐UTP-labeled tRNAphe were treated with recombinant TruB1, mt1, or mt2. The strong upper bands represent uridine (U), and the lower weaker bands represent pseudouridine (Ψ) on autoradiographs of the TLC plate.
Relative miRNA expression of let‐7a in HEK‐293 cells infected with tetracycline‐inducible expressing lentiviruses for TruB1, mt1, mt2, or GFP 5 days after doxycycline treatment, as determined by qRT–PCR. Significance was assessed using 2‐tailed Student's t‐test, < 0.05*.
Northern blotting for let‐7a and RNU6B in HeLa cells infected with lentiviruses encoding tetracycline‐inducible expression of TruB1, mt1, mt2, or GFP, 5 days after doxycycline treatment.
Hybridization intensities of (D) were quantified and normalized to ctrl (GFP). Significance was assessed using 2‐tailed Student's t‐test, < 0.05*.
Pseudouridylation activity of TruB1 for tRNA and pri‐miRNAs. 32p‐UTP-labeled tRNAphe, pri‐let-7a1, or pri‐miR-10a were treated with recombinant TruB1. Upper bands represent uridine (U), lower bands represent pseudouridine (Ψ) in autoradiographs of the TLC plate.
Location of pseudouridine sites detected by the CMC primer extension method. Total RNA purified from HEK‐293FT cells were treated with CMC. CMC‐treated RNA were reverse‐transcribed with RI‐labeled specific primers for tRNAphe or pri‐let-7b. ddATP was used for sequence control. Pseudouridines are indicated by black arrows.
Data information: All experiments were performed in triplicate. Error bars show SD.