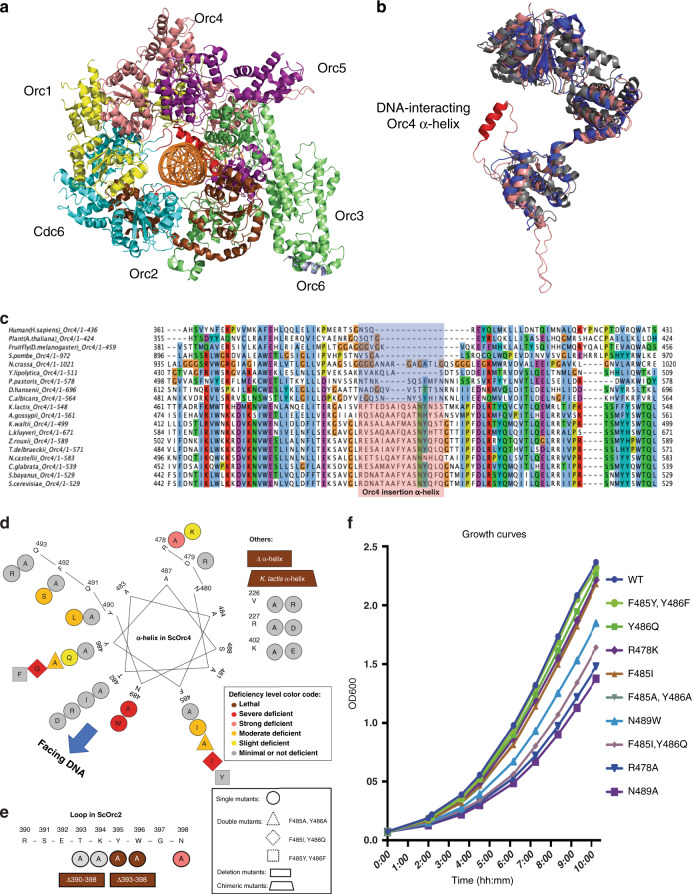
Fig. 1. DNA interacting Orc4 α-helix and Orc2 loop are essential.
a Top-view of ORC–Cdc6 structure encircling an origin DNA with Orc1–6 and Cdc6 indicated. Recolored from previous cryo-EM work6 OCCM structure (PDB code 5udb) and Orc4 α-helix and Orc2 loop that interact with DNA are colored in red. b Orc4 structure superposition of human Orc4 in blue (from PDB code 5uj7), Drosophila Orc4 in gray (from PDB code 4xgc) and S. cerevisiae Orc4 in salmon (from PDB code 5udb). Orc4 α-helix that interacts with DNA is colored in red. c Multiple sequence alignment of Orc4 among representing eukaryotic species as indicated. Orc4 α-helix region indicated with species that do not have sequence-specific origins shadowed in blue and species that sequence-specific origins exist shadowed in pink. d, e Maps of mutant viability phenotypes from the plasmid shuffle assay: Orc4 α-helix in a helical wheel d and Orc2 loop in connected line e. Mutant deficiency phenotypes (Supplementary Figs. 2 and 3) are summarized in color codes as indicated. Amino acids indicated in one-letter abbreviation. Different mutant types are indicated with different shapes. f Growth curves of NTAP-Orc4 integrated strains (see “Methods” section) in YPD with initiation OD600 at 0.05 at 30 °C to measure OD600 at different time points.