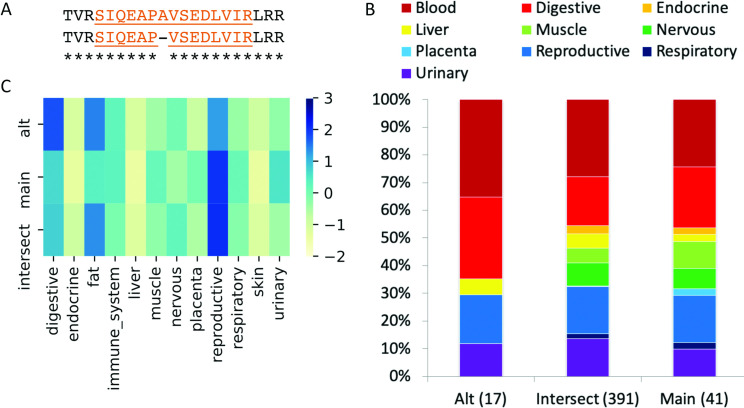
Fig 8. Group-specific splicing in gene TOR1AIP1.
(A) We found peptide evidence for a single primate-derived splice event for TOR1AIP1. This NAGNAG splicing event resulted in the loss/gain of a single amino acid. The discriminating peptides we detected are highlighted. (B) The distribution of the PEDs for the discriminating peptides (“Alt” and “Main”) and the remaining peptides that mapped to TOR1AIP1, but that did not distinguish one isoform from the other (“Intersect”). The number of PEDs in each set is shown in brackets. Fisher tests show that the distributions of PEDs between the Main and Alt peptides are not significantly different over any of the ten tissue groups. (C) The group-specific distribution of reads that support each side of the splice junction (main, alt) and those that support remaining common protein sequence (intersect) coloured by standard deviation from the mean; the darker the colour, the greater the positive standard deviation. There is more than one standard deviation between the reads for the digestive and reproductive groups, so the TOR1AIP1 event is determined to be group specific at the transcript level for these tissue groups.