Figure 5.
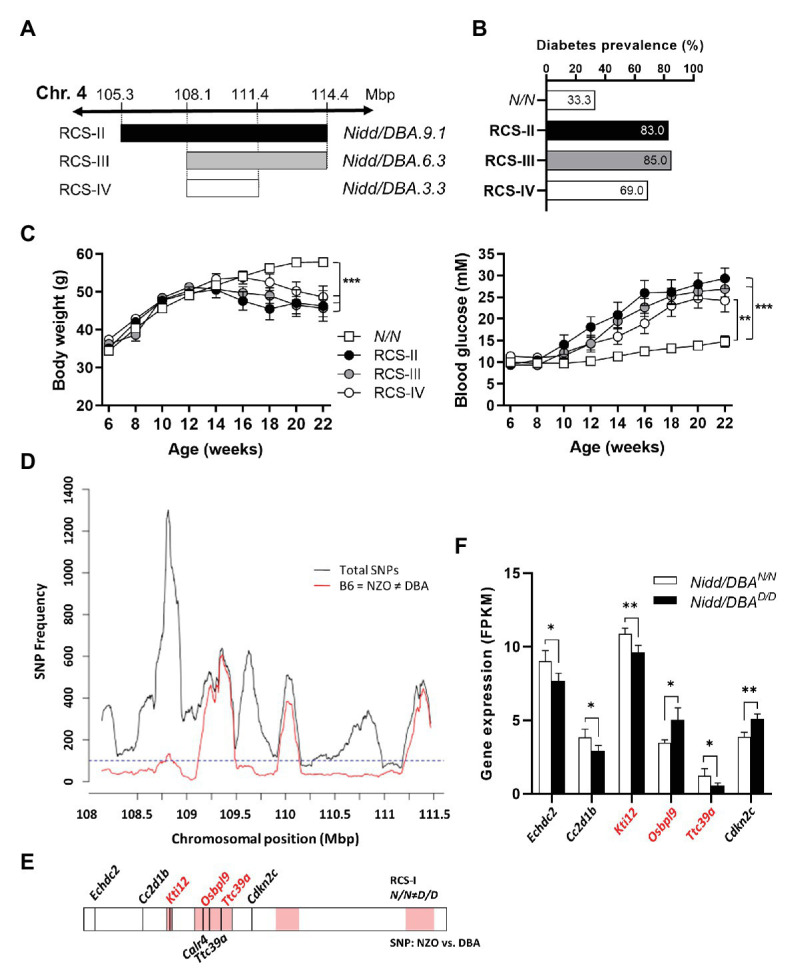
Combined approach of haplotype and gene expression analysis in islets for the identification of DBA-specific gene variants within the Nidd/DBA locus. (A) Generation of congenic mice carrying different fragments of the QTL Nidd/DBA. (B) Diabetes prevalence (blood glucose >16.6 mM, week 16), (C) body weight, and blood glucose development in congenic mice carrying one heterozygous DBA allele for the Nidd/DBA locus compared to homozygous NZO controls. (D) Haplotype map of single nucleotide polymorphisms (SNPs) within the critical region of Nidd/DBA (108.8–111.4 Mbp). Black line: total number of SNPs (all SNPs annotated for the B6 reference genome with calls for NZO and DBA). Red line: SNPs according to B6 = NZO ≠ DBA. (E) QTL was dissected into windows of 250 kb. Red boxes indicate 100 SNPs per window polymorphic according to B6 = NZO ≠ DBA. Location of genes within the critical Nidd/DBA interval with a differential expression in pancreatic islets of 6 weeks old congenic mice (N/N ≠ D/D) and position of genes with non-synonymous coding variants between the parental strains NZO and DBA (REL-1505-GRCm38). (F) Genes within the critical Nidd/DBA interval displaying a differential expression in pancreatic islets of congenic mice (RCS-I, Nidd/DBA.13.6). Expression was analyzed via RNA-sequencing (n = 4). Genes with differential expression in pancreatic islets and location within a polymorphic Nidd/DBA haplotype block are indicated in red. Data are presented as means ± SEM and were analyzed with Student’s t-test (F) or one-way ANOVA (C). *p < 0.05; **p < 0.01; ***p < 0.001.
