ABSTRACT
The Ras oncogene is notoriously difficult to target with specific therapeutics. Consequently, there is interest to better understand the Ras signaling pathways to identify potential targetable effectors. Recently, the mechanistic target of rapamycin complex 2 (mTORC2) was identified as an evolutionarily conserved Ras effector. mTORC2 regulates essential cellular processes, including metabolism, survival, growth, proliferation and migration. Moreover, increasing evidence implicate mTORC2 in oncogenesis. Little is known about the regulation of mTORC2 activity, but proposed mechanisms include a role for phosphatidylinositol (3,4,5)-trisphosphate – which is produced by class I phosphatidylinositol 3-kinases (PI3Ks), well-characterized Ras effectors. Therefore, the relationship between Ras, PI3K and mTORC2, in both normal physiology and cancer is unclear; moreover, seemingly conflicting observations have been reported. Here, we review the evidence on potential links between Ras, PI3K and mTORC2. Interestingly, data suggest that Ras and PI3K are both direct regulators of mTORC2 but that they act on distinct pools of mTORC2: Ras activates mTORC2 at the plasma membrane, whereas PI3K activates mTORC2 at intracellular compartments. Consequently, we propose a model to explain how Ras and PI3K can differentially regulate mTORC2, and highlight the diversity in the mechanisms of mTORC2 regulation, which appear to be determined by the stimulus, cell type, and the molecularly and spatially distinct mTORC2 pools.
KEY WORDS: Ras GTPase, Phosphatidylinositol 3-kinase, Mechanistic target of rapamycin complex 2
Summary: Current evidence suggests conserved but separate roles of Ras and PI3K in the direct activation of mTORC2, implicating the regulation of distinct cellular mTORC2 pools downstream of different stimuli.
Introduction
Ras is an evolutionarily conserved small GTPase and notorious oncogene (Pylayeva-Gupta et al., 2011; Simanshu et al., 2017). In mammalian cells, Ras is mostly characterized for its role in mediating the cellular response to growth factors, which leads to modulation of gene expression necessary for cell growth and proliferation through activation of the extracellular-regulated kinase (ERK) signaling cascade (Malumbres and Barbacid, 2003). The three closely related human Ras isoforms, H-Ras, N-Ras and K-Ras, have been extensively studied because of their involvement in tumorigenesis, with ∼25% of tumors expressing an active mutant form of one of these Ras proteins (Khan et al., 2020). Ras proteins act as molecular switches, cycling between GTP-bound active and GDP-bound inactive states. This activity cycle is regulated by (i) guanosine exchange factors (GEFs) that stimulate the exchange of GDP for GTP on Ras, thereby active Ras; and (ii) GTPase-activating proteins (GAPs) that stimulate the intrinsic GTPase activity of Ras, leading to hydrolysis of GTP into GDP and, thereby, inactivating Ras (Vigil et al., 2010; Nakhaei-Rad et al., 2018). Ras oncogenic mutations are those that result in a persistent GTP-bound, active state. The most common oncogenic Ras mutations are substitution of a single amino acid at positions 12, 13 or 61, which induces a constitutively active Ras phenotype (Muñoz-Maldonado et al., 2019). Apart from the K-Ras G12C mutation, with the cysteine offering a chemical handle for covalent inhibitors, oncogenic Ras has proven extremely hard to target directly (Ostrem et al., 2013; McCormick, 2019). Ras effectors and downstream pathways are also under consideration for potential cancer treatments and, therefore, a lot of interest and effort is placed into identifying new Ras effector pathways (Engin et al., 2017).
Well-described Ras effectors include the serine/threonine kinase Raf (Vojtek et al., 1993), class I phosphatidylinositol 3-kinases (PI3Ks; hereafter referred to as PI3K unless indicated otherwise) (Kodaki et al., 1994; Rodriguez-Viciana et al., 1994) and the ral guanine nucleotide dissociation stimulator RALGDS (Hofer et al., 1994). These, and many other known effectors of Ras, interact with the switch-I region of Ras through an ubiquitin-like fold structure, forming an interprotein beta-sheet (Wohlgemuth et al., 2005). These conserved structures have been termed Ras association (RA) or Ras binding (RB) domains, depending on the set of residues involved in the interaction (Wohlgemuth et al., 2005). Other identified effectors of Ras, such as Src (Thornton et al., 2003) and calmodulin (Abraham et al., 2009), do not interact through a canonical RA or RB domain. By using Dictyostelium discoideum as an experimental model system, we identified the mechanistic target of rapamycin complex 2 (mTORC2) to be a non-canonical effector of Ras (Khanna et al., 2016), which was recently found to be conserved in human cells (Kovalski et al., 2019; Lone et al., 2019) and to play a role in cancer (Kovalski et al., 2019).
mTORC2 is one of two multiprotein complexes formed by the mTOR serine/threonine kinase (Jhanwar-Uniyal et al., 2019). mTORC1 is a master regulator of cell growth, controlling metabolism, protein synthesis and autophagy (Liu and Sabatini, 2020), whereas mTORC2 is best characterized for its evolutionarily conserved roles in regulating cell survival and actin cytoskeleton reorganization (Xie et al., 2018). While much is known about the biochemistry and regulation of mTORC1 cellular functions, the mechanisms governing mTORC2 activation and function remain largely unknown. For detailed descriptions of the structure, pathways and cellular functions of the mTOR complexes, we refer to other recent Reviews on these general topics (Xie et al., 2018; Jhanwar-Uniyal et al., 2019; Liu and Sabatini, 2020). Previous work suggests that mTORC2 activation by growth factor stimulation is mediated by PI3K, a well-characterized Ras effector (Gan et al., 2011; Liu et al., 2015). Consequently, there is increasing interest to understand the relationship between Ras, mTORC2 and PI3K (Fig. 1). Here, we review the Ras-mediated regulation of PI3K and mTORC2, including work performed by using eukaryotic model organisms that contributed to our current knowledge of these pathways. We also discuss potential mechanisms and contexts in which the pathways may intersect.
Fig. 1.
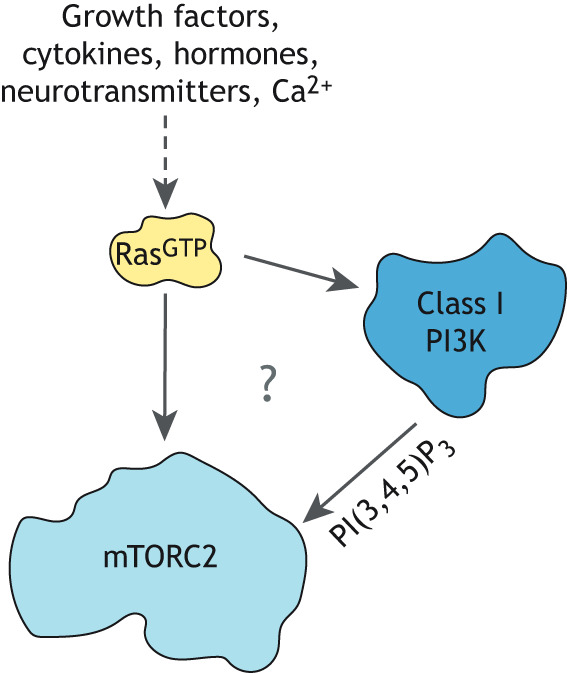
The unclear relationship between Ras, PI3K and mTORC2. Ras is activated by many different stimuli and its intracellular effectors include class 1 phosphatidylinositol 3-kinases (PI3Ks) that produce PI(3,4,5)P3 and, as recently identified, mTORC2. Several studies suggest that PI(3,4,5)P3 promotes mTORC2 activation. Whether Ras and PI3K cooperate to fully activate mTORC2 or whether they act independently to promote mTORC2 activation is still debated.
RAS regulation of PI3K
There are three different classes of PI3K, which synthesize three different phosphoinositides (Jean and Kiger, 2014). Class I PI3Ks are heterodimeric enzymes that synthesize the signaling lipid phosphatidylinositol (3,4,5)-trisphosphate [PI(3,4,5)P3] and have an RB domain in their p110 catalytic subunits. Only the class I subunits (PIK3CA, PIK3CD and PIK3CG; also known as to as PI3Kα, PI3Kδ and PI3Kγ, respectively) are stimulus-dependent Ras effectors (Vanhaesebroeck et al., 2010; Rathinaswamy and Burke, 2020). Ras-mediated activation of PI3K can occur downstream of a variety of signaling inputs, including receptor tyrosine kinases (RTKs) and seven transmembrane receptors [7TMs; also called G protein-coupled receptors (GPCRs)], and is conserved throughout evolution, as highlighted by the important role of this pathway in the primitive eukaryote and model organism Dictyostelium discoideum (Funamoto et al., 2001, 2002; Sasaki et al., 2004; 2007; Comer et al., 2005; Comer and Parent, 2006; Charest and Firtel, 2007; Takeda et al., 2007; Bosgraaf et al., 2008; Zhang et al., 2008; Gruver et al., 2008; Kölsch et al., 2008). The best-understood activation mechanism of class I PI3K is the one where it is directly activated by RTKs in mammalian cells: the Src homology 2 (SH2) domains of PI3K bind to phosphorylated tyrosine residues on an activated RTK, leading to a conformational change in the active site of PI3K, thereby stimulating its catalytic activity (Fruman et al., 2017). Whether the mechanism through which Ras regulates PI3K activity involves such an allosteric activation mechanism, resulting from Ras binding to the RB domain of PI3K, is still unclear. However, recent studies suggest that, at least for the RTK-induced activation of mammalian PIK3CA, Ras mostly acts to stabilize PI3K at the plasma membrane, whereas PI3K is allosterically activated by the RTK (Buckles et al., 2017; Nussinov et al., 2019).
Ras-mediated regulation of PI3K plays a role in many cellular processes involved in both normal physiology and disease (Castellano and Downward, 2011). In all of these contexts, the Ras-stimulated production of PI(3,4,5)P3 results in the membrane recruitment and, in many cases, activation of pleckstrin homology (PH) domain-containing proteins (Riehle et al., 2013). These PH domain-containing proteins include the well-studied 3-phosphoinositide-dependent protein kinase 1 (PDPK1, herafter referred to as PDK1) and AKT kinases, GEFs for Rac small GTPases and Bruton tyrosine kinase (Btk) (Fig. 2). The best-characterized roles of Ras–PI3K signaling in eukaryotes are the regulation of AKT function in cell survival and growth, as well as the remodeling of the actin cytoskeleton (Kölsch et al., 2008; Hoxhaj and Manning, 2019). In addition to promoting its recruitment to the plasma membrane, mammalian PI3K promotes activation of AKT through PI(3,4,5)P3-mediated recruitment to the membrane and activation of PDK1, which, in turn, activates AKT by phosphorylating threonine residue 308 within its activation loop (Stokoe et al., 1997; Vanhaesebroeck and Alessi, 2000). The Ras–PI3K pathway can also regulate the actin cytoskeleton by promoting Rac signaling in various cellular contexts (Campa et al., 2015).
Fig. 2.
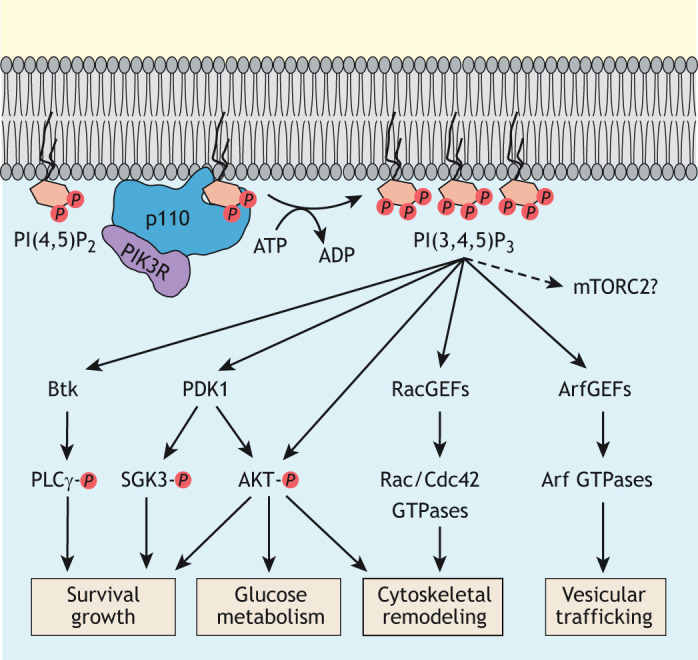
Class I PI3K signaling. Class 1 PI3Ks consist of a p110 catalytic subunit and a varying regulatory subunit (PIK3R). These PI3Ks are responsible for the production of the signaling lipid PI(3,4,5)P3, which can trigger many different intracellular signaling pathways involved in promotion of cellular survival, cell growth and glucose uptake, as well as in modulation of metabolism, actin cytoskeleton and vesicular trafficking. A role for mTORC2 downstream from PI3K is still debated. Btk, Bruton tyrosine kinase. PLCγ, phospholipase C gamma. SGK3, serum/glucocorticoid-regulated kinase-3. RacGEFs, Rac guanine exchange factors. ArfGEFs, Arf guanine exchange factors, PDK1, phosphoinositide-dependent kinase-1.
Work in Dictyostelium greatly contributed to what is currently known about the role of Ras–PI3K signaling in the control of the actin cytoskeleton. Ras, PI3K and actin in Dictyostelium are linked through a positive-feedback loop and control cell migration as well as cytokinesis (Sasaki et al., 2004, 2007; Janetopoulos et al., 2005; Arai et al., 2010). In this feedback loop, PI(3,4,5)P3-mediated polymerization of filamentous actin (F-actin) leads to increased Ras and PI3K activation; this results in localized amplification of the PI(3,4,5)P3 signal, which, in turn, leads to pseudopod protrusion in migrating cells (Sasaki et al., 2004, 2007; Arai et al., 2010) and to separation of the poles in dividing cells (Janetopoulos et al., 2005). Similar feedback loops that involve Ras, PI3K and actin also amplify PI3K signaling in mammalian cells (Peyrollier et al., 2000; Wang et al., 2002; Yang et al., 2012), and promote fibroblast migration (Thevathasan et al., 2013). Compelling evidence support the notion that Ras–PI3K signaling plays a conserved role in controlling the migration of many types of mammalian cell, including fibroblasts (Thevathasan et al., 2013; Castellano et al., 2016), neutrophils (Suire et al., 2006), smooth muscle cells (Tanski et al., 2002) and endothelial cells (Eller-Borges et al., 2015). In Dictyostelium cells that migrate in response to the chemoattractant cyclic AMP (cAMP), the Ras–PI3K pathway is primarily important to respond to shallow cAMP gradients, and to increase the speed and directional accuracy of migration in steeper cAMP gradients, which is likely to be attributable to the PI(3,4,5)P3 amplification feedback loop (Postma et al., 2004; Takeda et al., 2007; Bosgraaf et al., 2008; Gruver et al., 2008). Ras–PI3K signaling in migrating Dictyostelium cells also controls the pathway leading to the intracellular production and, then, secretion of cAMP (Comer et al., 2005; Comer and Parent, 2006). This pathway functions to relay the cAMP chemoattractant signal to other Dictyostelium cells, thereby attracting many cells for aggregate formation during development (Mahadeo and Parent, 2006). Interestingly, it is in the context of Dictyostelium chemotaxis and chemoattractant signal relay regulation that Ras was first discovered to also signal through mTORC2 (Lim et al., 2001; Lee et al., 2005; Bolourani et al., 2006; Kamimura et al., 2008; Cai et al., 2010; Charest et al., 2010).
RAS regulation of mTORC2
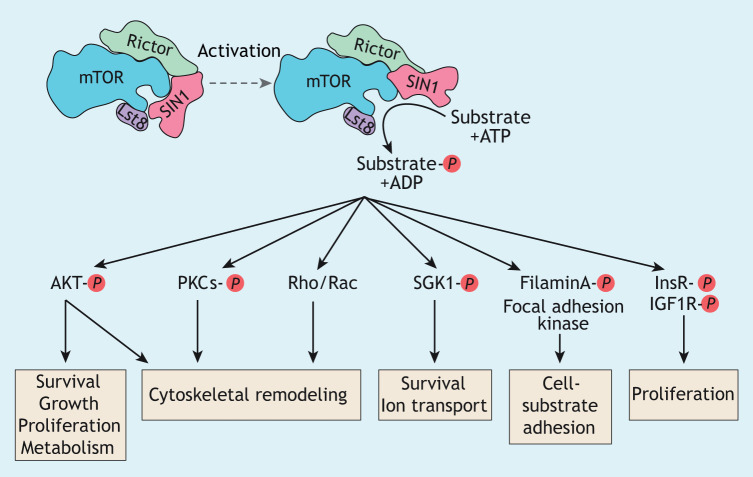
mTORC2 plays evolutionarily conserved roles, controlling the actin cytoskeleton and cell survival (Xie et al., 2018). In yeast, mTORC2 also acts as a sensor to control lipid and protein composition of the plasma membrane (Roelants et al., 2017). In Dictyostelium, mTORC2 controls phagocytosis and is central to the directed migration of cells (chemotaxis) in response to the chemoattractant cAMP by regulating the actin cytoskeleton and cAMP synthesis (Lee et al., 2005; Charest et al., 2010; Kamimura and Devreotes, 2010; Rosel et al., 2012). In mammalian cells, mTORC2 controls the actin cytoskeleton and cell migration in addition to several other cellular functions, such as ion transport, cell-substrate adhesion, cellular survival, proliferation and growth, as well as metabolism (Fig. 3) (Jacinto et al., 2004; Zhou and Huang, 2010; Liu et al., 2010; Gulhati et al., 2011; Farhan et al., 2015; Chen et al., 2015a,b; Yin et al., 2016; Sato et al., 2016; Roelants et al., 2017; Xie et al., 2018; Jaiswal et al., 2019; Liu and Sabatini, 2020). Similar to that in Dictyostelium, mTORC2-mediated regulation of the actin cytoskeleton in mammalian cells is involved in controlling the motility, migration and chemotaxis of many types of cell, including neutrophils and human cancer cells (Zhou and Huang, 2010; Gulhati et al., 2011; Liu and Parent, 2011; Kim et al., 2017; Holroyd and Michie, 2018; Liu and Sabatini, 2020). Many of the known functions of mTORC2 in human cells are linked to its direct regulation of AKT activity through phosphorylation of serine residue 473 in its C-terminal hydrophobic motif, which, together with PDK1-mediated activation loop phosphorylation, results in full activation of AKT (Sarbassov et al., 2005; Hoxhaj and Manning, 2019). Other known substrates of mammalian mTORC2 include protein kinase C (PKC), serum/glucocorticoid-regulated kinase 1 (SGK1), filamin A, the insulin receptor (InsR) and the insulin growth factor 1 receptor (IGF1R) (Fig. 3) (Chantaravisoot et al., 2015; Sato et al., 2016; Yin et al., 2016; Liu and Sabatini, 2020).
Fig. 3.
mTORC2 signaling. Shown are the evolutionarily conserved components of mTORC2, with SIN1 occluding the active site of mTOR. Activation of mTORC2 probably involves displacement of SIN1 from the active site of mTOR, thereby allowing access to the substrates and their phosphorylation. Known mTORC2 substrates and regulated cellular pathways and processes are also shown. IGF1R, insulin-like growth factor1 receptor; PKCs, protein kinase C family members.
In a yeast-two-hybrid screen for Dictyostelium proteins that interact with the active form of mammalian H-Ras (Lee et al., 1999), one of the identified Ras-interacting proteins, i.e. Ras interacting protein 3 (RIP3), was later found to be the homologue of mammalian mTORC2 subunit MAPKAP1 (also known and hereafter referred to as SIN1), a unique and essential component of mTORC2 (Myers et al., 2005; Jacinto et al., 2006; Yang et al., 2006). SIN1 and the rapamycin-insensitive companion of mTOR (RICTOR) are two evolutionarily conserved unique components of mTORC2 that are essential for its integrity and provide functional specificity, whereas the other conserved subunits mTOR and lethal with SEC13 protein 8 (LST8) are part of both mTOR complexes (Liu and Sabatini, 2020) (Fig. 3). SIN1, which has RB and PH domains, is linked to the recognition of mTORC2 substrates as well as the regulation of its localization and activation (Jacinto et al., 2006; Schroder et al., 2007; Bracho-Valdes et al., 2011; Cameron et al., 2011; Liu et al., 2015; Tatebe et al., 2017; Yao et al., 2017). Five human SIN1 isoforms have been described but only SIN1.1, SIN1.2 and SIN1.5 – the latter being a much smaller isoform that lacks most of the N- and C-terminal domains – have been found to assemble into mTORC2 and form three distinct complexes, with only SIN1.1- and SIN1.2-containing mTORC2 regulated by insulin stimulation (Frias et al., 2006). Recent insights from high resolution cryo-EM structures of mTORC2, together with those from previous biochemical studies, revealed that SIN1 interacts with the FK506-binding protein 1b (FKBP12)-rapamycin-binding (FRB) and kinase domains of mTOR; this occludes the active site of mTOR, thereby inhibiting mTORC2 while, nevertheless, having a role in substrate recruitment (Jacinto et al., 2006; Liu et al., 2015; Karuppasamy et al., 2017; Tatebe et al., 2017; Chen et al., 2018; Stuttfeld et al., 2018). Consequently, it is generally assumed that any mTORC2 activation mechanism includes a conformational change of SIN1 in order to free the active site of mTOR (Fig. 3).
Interestingly, although the Dictyostelium SIN1 homologue RIP3 (officially known as RipA) interacts in vitro with H-Ras and the Dictyostelium Ras protein RasG, in vivo studies revealed that the Ras protein RasC is responsible to promote mTORC2 activation (Kamimura et al., 2008; Cai et al., 2010; Charest et al., 2010). Active RasC directly binds the kinase domain of mTOR and not the RB domain of RIP3 (Khanna et al., 2016). However, the Ras-related protein Rap1 binds to the RB domain of RIP3 and regulates the RasC-mediated activation of mTORC2 (Khanna et al., 2016). Furthermore, in its GDP-bound form, the Dictyostelium RhoA GTPase RacE, was also recently found to play a role in promoting activation of mTORC2 by associating with RasC (Senoo et al., 2019). Notably, human Rap1b was also found to bind the RB domain of human SIN1 in a GTP-dependent manner in vitro (Khanna et al., 2016), and human RhoA was also shown to pull down mTORC2 in human embryonic kidney 293(HEK293) cells (Senoo et al., 2019). Therefore, these observations suggest that both a Rap1- and human RhoA-mediated regulation of mTORC2 activity is conserved in mammalian cells, but further studies are needed to understand their biological implications. Interestingly, the small GTPase Rac1 also regulates mammalian mTORC2 by promoting its recruitment to the plasma membrane through a nucleotide-independent interaction with mTOR (Saci et al., 2011).
In mammalian cells, diverse stimuli and receptor systems can signal through mTORC2. The evidence suggests that the mechanism varies with the nature of cells and stimulus, contributing to the specificity of the response (Knudsen et al., 2020). For instance, the activity of mammalian mTORC2 was reported to be stimulated by the association of mTORC2 with ribosomes in a PI3K-dependent manner (Zinzalla et al., 2011), by cAMP signaling (Sato et al., 2014; Mukaida et al., 2017), release and/or degradation of DEP domain-containing mTOR-interacting protein (DEPTOR) (Peterson et al., 2009), phosphatidic acid binding to mTOR (Foster, 2013; Menon et al., 2017), association with PKC (Partovian et al., 2008; Gleason et al., 2019), Rac1-mediated recruitment to the plasma membrane (Saci et al., 2011), the AKT-mediated phosphorylation of SIN1 (Yang et al., 2015), binding of SIN1 to PI(3,4,5)P3 (Gan et al., 2011; Liu et al., 2015) and, as discussed here, direct interaction of Ras with mTORC2 (Khanna et al., 2016; Kovalski et al., 2019; Lone et al., 2019). Indeed, recent studies using oncogenic Ras mutants revealed that the binding of active Ras to mTORC2, which results in stimulation of mTOR activity, is conserved in human cells and, importantly, that this signaling pathway likely plays a general role in oncogenesis (Kovalski et al., 2019; Lone et al., 2019). In particular, a recent study that elegantly combined proximity-dependent proteomics and CRISPR genetics followed by rigorous in vitro and in vivo studies, identified mTORC2 as an effector of oncogenic H-Ras, N-Ras and K-Ras, which involves their direct interaction with mTOR and SIN1 (Kovalski et al., 2019). Results of this study further show that mTOR primarily interacts with oncogenic Ras through its kinase domain – as was found for Dictyostelium RasC (Khanna et al., 2016) – and with possible secondary sites located in the N-terminal HEAT domain region of mTOR, reminiscent of the binding of Rheb to mTOR in mTORC1 (Long et al., 2005; Avruch et al., 2009; Yang et al., 2017). In addition to binding mTOR, oncogenic Ras was also found to interact with the RB domain of SIN1 and to be crucial for Ras in order to interact with and activate mTORC2 (Kovalski et al., 2019). Consequently, as human Ras has previously been suggested to form dimers (Zhou et al., 2018), it was proposed that a Ras dimer binds mTORC2, with one Ras protomer interacting with the mTOR kinase domain while the other interacts with SIN1 (Kovalski et al., 2019).
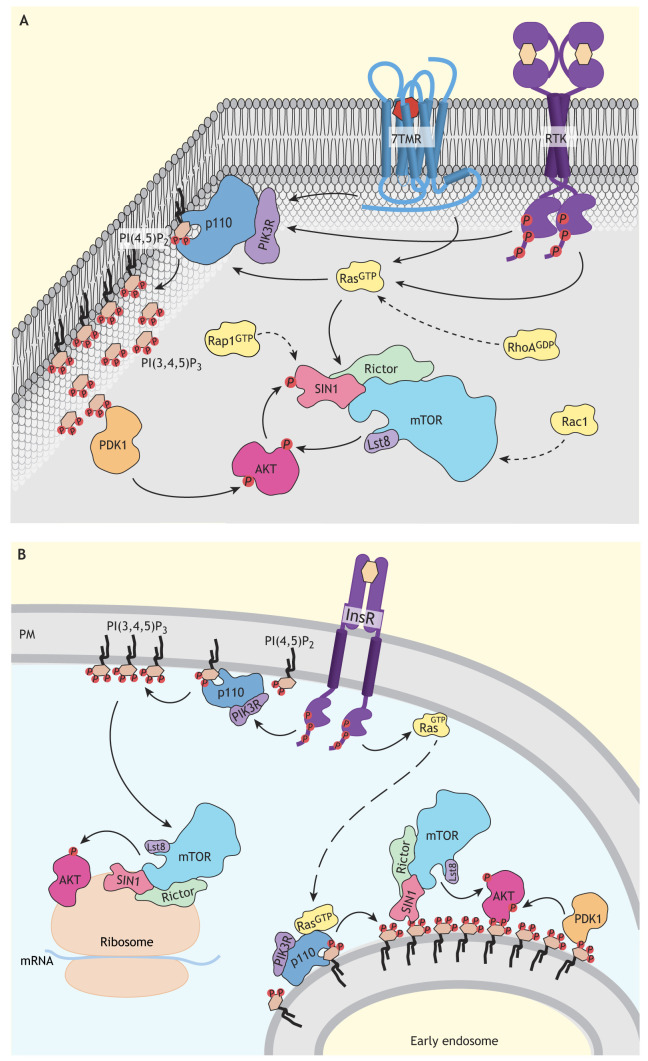
Considering the evidence and all reported observations with regard to Ras-mediated activation of mTORC2 in eukaryotes, we propose the idea of a general, conserved mechanism where Ras binds to the mTOR kinase domain, while another Ras family protein – whether an additional Ras, Rap1 or RhoA – simultaneously binds to the RB domain of SIN1 to fully activate mTORC2 (Fig. 4A). Therefore, the binding of one of the small GTPases Ras, Rap1 or RhoA to SIN1 could play two simultaneous roles: (1) the recruitment of mTORC2 to the plasma membrane; and (2) the displacement of SIN1 from the active site of mTOR, thereby assisting the Ras-mediated stimulation of kinase activity by direct binding of Ras to mTOR. Most likely, depending on the cells, stimulus or the intracellular localization of mTORC2, the mechanism to displace SIN1 and release its inhibition of mTOR kinase will vary. For example, instead of a Ras family protein binding to SIN1, its displacement from the active site of mTOR can be achieved by binding of PI(3,4,5)P3 to the PH domain of SIN1 (Gan et al., 2011; Liu et al., 2015).
Fig. 4.
Model of how Ras and PI3K might regulate distinct mTORC2 pools. (A) Illustration of proposed mechanisms regulating the mTORC2 pool at the plasma membrane. Several receptor systems, including seven transmembrane receptors (7TMRs) and receptor tyrosine kinases (RTKs), promote mTORC2 activation at the plasma membrane. Here, mTORC2 activity might primarily be stimulated by Ras binding to mTOR and/or SIN1. Additional small GTPases, including Rap1, RhoA and Rac1, probably contribute to activation of the mTORC2 pool at the plasma membrane. mTORC2 signaling at the plasma membrane includes activation of AKT, which is also regulated by PI3K-PDK1 signaling in human cells and can involve AKT-stimulated activation of mTORC2 through SIN1 phosphorylation in a positive feedback loop. (B) Illustration of proposed mechanisms regulating the intracellular mTORC2 pool. The insulin receptor (InsR) is likely to signal through PI3K to promote activation of intracellular mTORC2 pools, including at the ribosome and early endosomes. At the ribosome, active mTORC2 phosphorylates AKT and other AGC kinases within their turn motif, which contributes to the folding and stability of these kinases. PI3K-mediated activation of mTORC2 at early endosomes could occur downstream of Ras and results in binding of the SIN1 PH domain to PI(3,4,5)P3, thereby freeing the active site of mTOR. mTORC2 located at early endosomes signals through AKT, which is also likely to be regulated by phosphoinositide-dependent kinase-1 (PDK1) at this site. It is also possible that AKT amplifies the activity of mTORC2 as well as its own activity through a positive feedback loop involving SIN1 phosphorylation at early endosomes (not shown). The resulting activation of different cellular pools of AKT and, probably, also other localized mTORC2 substrates (not shown), is expected to play a crucial role in mediating a cellular response that is tailored to each stimulus. PIK3R, PI3K regulatory subunit.
PI3K regulation of mTORC2
Evidence suggests that PI3K promotes mTORC2 activation in mammalian cells, either directly through binding of PI3K-produced PI(3,4,5)P3 to SIN1 (Gan et al., 2011; Liu et al., 2015) or indirectly as part of pathways that lead to mTORC2 activation (Zinzalla et al., 2011; Liu et al., 2013; Yang et al., 2015). Such indirect PI3K regulation of mTORC2 includes the insulin-induced PI3K signaling-dependent association of mTORC2 with ribosomes, an interaction proposed to induce mTORC2 activation (Zinzalla et al., 2011). At the ribosome, mTORC2 is then positioned to co-translationally phosphorylate members of the protein kinase A, PKG and PKC (AGC) kinase family at their turn motif, found in some AGC kinases downstream of the catalytic domain, which is crucial for the folding and stability of AKT and conventional PKC (cPKC) (Facchinetti et al., 2008; Ikenoue et al., 2008; Oh et al., 2010). Therefore, in this context, the regulation of mTORC2 activity downstream of insulin-PI3K signaling provides a mechanism to couple insulin-induced protein synthesis and quality control. Another indirect mechanism for the PI3K-mediated regulation of mTORC2 in mammalian cells is the PI(3,4,5)P3- and PDK1-dependent, AKT-mediated phosphorylation of SIN1 on threonine residue 86 (T86) as part of a positive feedback loop (Liu et al., 2013; Yang et al., 2015). In response to growth factor stimulation, PDK1-mediated phosphorylation of the AKT activation loop first increases the kinase activity of AKT, which then phosphorylates SIN1, leading to enhanced mTORC2 kinase activity; following that, mTORC2 phosphorylates the hydrophobic motif of AKT, thereby achieving full AKT activation (Fig. 4A). The exact mechanism through which mTORC2 is activated upon T86 phosphorylation of SIN1 remains unknown; however, it was proposed to involve a weakening of the interaction between SIN1 and mTOR within mTORC2, as concurrent phosphorylation of T86 and T398 of SIN1 promotes its complete dissociation from the complex (Liu et al., 2013).
The PH domain of human SIN1 binds phosphatidylinositol and its phosphorylated derivatives as well as phosphatidic acid (PA) with no clear preference in lipid overlays, but it preferentially binds to PI(3,4,5)P3 in more physiologically-relevant liposomes (Schroder et al., 2007; Liu et al., 2015). The binding of SIN1 to PI(3,4,5)P3 appears highly conserved because the PH domain of Avo1, the yeast homologue of mammalian SIN1, was also found to bind phosphatidylinositol (4,5)-bisphosphate [PI(4,5)P2], i.e. the yeast functional homologue of PI(3,4,5)P3 in higher eukaryotes (Strahl and Thorner, 2007; Berchtold and Walther, 2009). The role of Avo1/SIN1 when binding to PI(4,5)P2/PI(3,4,5)P3, however, is still debated. It has initially been proposed that AvoI/SIN1 binding to PI(4,5)P2/PI(3,4,5)P3 represents an evolutionarily conserved role of this mTORC2 subunit in promoting the localization of mTORC2 to the plasma membrane. In yeast, mTORC2, indeed, partly localizes to membranes, particularly within discreet actin-independent foci at the plasma membrane (Kunz et al., 2000; Wedaman et al., 2003; Sturgill et al., 2008; Berchtold and Walther, 2009; Martinez Marshall et al., 2019). However, recent evidence suggests that the association of yeast Avo1 with the plasma membrane is independent of its PH domain and PI(4,5)P2. Instead, through its Armadillo repeats, Avo3 – the yeast homologue of mammalian RICTOR – promotes localization of mTORC2 at the plasma membrane (Martinez Marshall et al., 2019). To our knowledge, a role for RICTOR in targeting mammalian mTORC2 to membranes has yet to be explored. Nevertheless, there is strong evidence suggesting that the PH domain of mammalian SIN1 mediates its localization to both intracellular membranes and the plasma membrane, and that mTORC2 complexes formed with the PH domain-containing SIN1 isoforms SIN1.1 and SIN1.2 also display this localization (Frias et al., 2006; Schroder et al., 2007; Liu et al., 2015; Ebner et al., 2017). Intriguingly though, whereas the PH domain of human SIN1 – when expressed alone in cells – mostly localizes to the cytosol and translocates to the plasma membrane in response to insulin stimulation (Liu et al., 2015), full-length SIN1 is already partly located at the plasma membrane in resting cells and does not relocalize in response to insulin stimulation (Schroder et al., 2007; Ebner et al., 2017). Therefore, despite the fact that the PH domain of human SIN1 binds PI(3,4,5)P3 and is necessary for its plasma membrane localization, together, the evidence suggests a non-causal relationship.
Other evidence points to PI(3,4,5)P3 in directly stimulating mTORC2 activity. As previously mentioned, recent structural studies of both yeast and human mTORC2 revealed that Avo1/SIN1 interacts with the mTOR kinase domain in a way that occludes the active site of the latter (Karuppasamy et al., 2017; Chen et al., 2018; Stuttfeld et al., 2018). Furthermore, a previous biochemical study using human cells showed that the interaction of SIN1 with the kinase domain of mTOR involves the PH domain of SIN1 and, possibly, its N-terminal domain, thereby inhibiting mTORC2 activity towards AKT (Liu et al., 2015). Further observations in this study indicate that binding of PI(3,4,5)P3 to the PH domain of SIN1 can relieve inhibition of mTOR kinase by SIN1, leading to activation of mTOR kinase (Liu et al., 2015). Interestingly, the PH domain of yeast Avo1 was also found to be necessary for mTORC2 activity, although it is unknown whether binding of PI(4,5)P2 has a role in promoting activation (Martinez Marshall et al., 2019). In fact, the PI(3,4,5)P3-induced mTORC2 activation might not be universal because Dictyostelium strains that lack PI3K activity display normal mTORC2 activity and function during chemotaxis (Kamimura et al., 2008). Furthermore, a direct activation of human mTORC2 by PI(3,4,5)P3 is still debated. Although two studies reported that PI(3,4,5)P3 stimulates activation of purified mTORC2 in vitro (Gan et al., 2011; Liu et al., 2015), another study, using a very similar experiment, observed no effect of PI(3,4,5)P3 on mTORC2 activity (Frias et al., 2006). The main difference between these studies is the nature of the liposomes used to carry PI(3,4,5)P3; thus, it would be interesting to evaluate the effect of the other lipids included in the liposomes on mTORC2 activity.
The use of LocaTOR2, a cleverly designed compartment-specific biochemical reporter of mTORC2 activity that uses AKT phosphorylation as readout, provided valuable insight to the intracellular locations in which human mTORC2 is activated and to its regulation by PI(3,4,5)P3 (Ebner et al., 2017). In this regard, although the PH domain of SIN1 was found to be essential for the plasma membrane localization of mTORC2, observations suggest that this localization as well as the activity of mTORC2 at this site are not regulated by PI(3,4,5)P3 (Ebner et al., 2017). Instead, PI3K signaling was found to promote activation of an endosomal pool of mTORC2 (Ebner et al., 2017). Moreover, since recruitment of AKT to the plasma membrane automatically leads to phosphorylation of its hydrophobic motif in human cells, it was proposed that the plasma membrane pool of human mTORC2 is constitutively active (Ebner et al., 2017). However, whether SIN1 in mTORC2 at the plasma membrane of the human cells used in this study binds to other lipids or small GTPases that displace it from the active site of mTOR to promote mTORC2 activation remains to be determined.
mTORC2 activation – RAS and/or PI3K?
As the LocaTOR2 reporter revealed, the localization of mTORC2 in mammalian cells is likely to play a key role in determining its mechanism of activation and regulation, as well as, most likely, its cellular function (Ebner et al., 2017). Interestingly, components of mTORC2 were found to localize, in addition to the plasma membrane, to the cytosol, mitochondria, endoplasmic reticulum, endosomes, lysosome, Golgi, nucleus, perinuclear region and with ribosomes (reviewed in Knudsen et al., 2020). However, since RICTOR and SIN1 have been shown to play cellular roles that are independent of mTORC2 (Oh and Jacinto, 2011; He et al., 2013; Smrz et al., 2014; Gkountakos et al., 2018), an individual evaluation of their localization does not necessarily reflect the localization of mTORC2. We propose that mTORC2 is probably differentially regulated by Ras and PI3K, depending on its subcellular localization, which could also be influenced by the presence of the different SIN1 isoforms in the complex (Frias et al., 2006; Ebner et al., 2017). In fact, evidence from both Dictyostelium and mammalian cells suggest that the plasma membrane localization of mTORC2 is not regulated by PI3K (Kamimura et al., 2008; Ebner et al., 2017) (Fig. 4A).
Interestingly, only the plasma membrane pool of mTORC2 was found to be activated by oncogenic Ras (Kovalski et al., 2019). Although it remains to be determined whether wild-type Ras promotes mTORC2 activation at the plasma membrane the same way as oncogenic Ras, current evidence supports a model in which direct Ras-mediated activation of mTORC2 occurs at the plasma membrane and independently of PI3K (Kovalski et al., 2019; Lone et al., 2019) (Fig. 4A). In this model, PI3K still plays a role in the full activation of AKT at the plasma membrane of human cells by directing PDK1-mediated phosphorylation of the AKT activation loop; moreover, AKT-mediated phosphorylation of SIN1 may also contribute to full AKT activation through positive feedback. We further propose that another small GTPase, i.e. Rap1, RhoA or Rac1, also contributes to the activation of the mTORC2 pool at the plasma membrane; however, this is likely to be dictated by context-dependent conditions that have yet to be determined. In contrast, evidence points to PI3K primarily promoting the activation of the mTORC2 endosomal pool, as well as mTORC2 at the ribosome (Gan et al., 2011; Zinzalla et al., 2011; Ebner et al., 2017). The insulin-induced PI3K-mediated activation of mTORC2 at early endosomes in human cells might involve Ras, but in a pathway in which Ras acts upstream of PI3K (Gan et al., 2011) (Fig. 4B).
Conclusions and perspectives
Currently, all the evidence points to separate roles of Ras and PI3K regarding the direct activation of mTORC2: Ras binds and activates mTORC2 at the plasma membrane, and PI3K – through PI(3,4,5)P3 – activates the intracellular pools of mTORC2. It is important to consider, though, that most of the existing data on mTORC2 activity regulation in mammalian cells come from studies that used insulin as stimulus and phosphorylation of the AKT hydrophobic motif as readout. However, compelling evidence indicate that the regulation and signaling specificity of mTORC2 varies with the nature of the extracellular stimulus. For example, mTORC2 was found to be activated by PKC downstream of both syndecan-4 and the angiotensin II receptor type 1 (AGTR1), but resulting in completely different responses (Partovian et al., 2008; Gleason et al., 2019). Syndecan-4, which can act as a growth factor receptor or co-receptor, promotes mTORC2 activity towards AKT at the plasma membrane in response to fibroblast growth factor 2 (FGF2), insulin growth factor 1 (IGF1) and vascular endothelial growth factor (VEGF) through PKC (Partovian et al., 2008). In contrast, the angiotensin II (AngII)-mediated stimulation of AGTR1 mediates mTORC2 activation in a PKC-dependent manner at a perinuclear region but not the plasma membrane, where mTORC2 selectively phosphorylates and activates serum/glucocorticoid regulated kinase 1 (SGK1) but not AKT (Gleason et al., 2019). Together with the findings that mTORC2 exists as different cellular pools, these findings illustrate the diversity of the mechanisms of mTORC2 regulation that are dictated by the stimulus as well as the cellular conditions. Whether Ras-mediated activation of mTORC2 at the plasma membrane and that induced by PI3K at endosomes are universal mechanisms used to control these distinct cellular mTORC2 pools, or whether these mechanisms are at play only under defined circumstances and sometimes cooperate to regulate the same mTORC2 pool remains to be explored. Given the increasing evidence implicating mTORC2 in cancer (Zhou and Huang, 2010; Zou et al., 2016; Kim et al., 2017; Gkountakos et al., 2018; Butt et al., 2019) and that it can be activated by the most commonly mutated oncogene Ras, a better understanding of the mechanisms that regulate mTORC2 and how it is linked to other Ras pathways will help in guiding the choice of increasingly personalized therapeutic anti-cancer strategies.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
This research received funding from the National Institutes of Health (grant number: 5R01GM131200-02). Deposited in PMC for release after 12 months.
References
- Abraham S. J., Nolet R. P., Calvert R. J., Anderson L. M. and Gaponenko V. (2009). The hypervariable region of K-Ras4B is responsible for its specific interactions with calmodulin. Biochemistry 48, 7575-7583. 10.1021/bi900769j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai Y., Shibata T., Matsuoka S., Sato M. J., Yanagida T. and Ueda M. (2010). Self-organization of the phosphatidylinositol lipids signaling system for random cell migration. Proc. Natl. Acad. Sci. USA 107, 12399-12404. 10.1073/pnas.0908278107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avruch J., Long X., Lin Y., Ortiz-Vega S., Rapley J., Papageorgiou A., Oshiro N. and Kikkawa U. (2009). Activation of mTORC1 in two steps: Rheb-GTP activation of catalytic function and increased binding of substrates to raptor. Biochem. Soc. Trans. 37, 223-226. 10.1042/BST0370223 [DOI] [PubMed] [Google Scholar]
- Berchtold D. and Walther T. C. (2009). TORC2 plasma membrane localization is essential for cell viability and restricted to a distinct domain. Mol. Biol. Cell 20, 1565-1575. 10.1091/mbc.e08-10-1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolourani P., Spiegelman G. B. and Weeks G. (2006). Delineation of the roles played by RasG and RasC in cAMP-dependent signal transduction during the early development of Dictyostelium discoideum. Mol. Biol. Cell 17, 4543-4550. 10.1091/mbc.e05-11-1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosgraaf L., Keizer-Gunnink I. and Van Haastert P. J. (2008). PI3-kinase signaling contributes to orientation in shallow gradients and enhances speed in steep chemoattractant gradients. J. Cell Sci. 121, 3589-3597. 10.1242/jcs.031781 [DOI] [PubMed] [Google Scholar]
- Bracho-Valdes I., Moreno-Alvarez P., Valencia-Martinez I., Robles-Molina E., Chavez-Vargas L. and Vazquez-Prado J. (2011). mTORC1- and mTORC2-interacting proteins keep their multifunctional partners focused. IUBMB Life 63, 880-898. 10.1002/iub.558 [DOI] [PubMed] [Google Scholar]
- Buckles T. C., Ziemba B. P., Masson G. R., Williams R. L. and Falke J. J. (2017). Single-molecule study reveals how receptor and Ras synergistically activate PI3Kα and PIP3 signaling. Biophys. J. 113, 2396-2405. 10.1016/j.bpj.2017.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt G., Shahwar D., Qureshi M. Z., Attar R., Akram M., Birinci Y., Karatoprak G. S., Gasparri M. L. and Farooqi A. A. (2019). Role of mTORC1 and mTORC2 in breast cancer: therapeutic targeting of mTOR and its partners to overcome metastasis and drug resistance. Adv. Exp. Med. Biol. 1152, 283-292. 10.1007/978-3-030-20301-6_15 [DOI] [PubMed] [Google Scholar]
- Cai H., Das S., Kamimura Y., Long Y., Parent C. A. and Devreotes P. N. (2010). Ras-mediated activation of the TORC2-PKB pathway is critical for chemotaxis. J. Cell Biol. 190, 233-245. 10.1083/jcb.201001129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron A. J. M., Linch M. D., Saurin A. T., Escribano C. and Parker P. J. (2011). mTORC2 targets AGC kinases through Sin1-dependent recruitment. Biochem. J. 439, 287-297. 10.1042/BJ20110678 [DOI] [PubMed] [Google Scholar]
- Campa C. C., Ciraolo E., Ghigo A., Germena G. and Hirsch E. (2015). Crossroads of PI3K and Rac pathways. Small GTPases 6, 71-80. 10.4161/21541248.2014.989789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano E. and Downward J. (2011). RAS interaction with PI3K: more than just another effector pathway. Genes Cancer 2, 261-274. 10.1177/1947601911408079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano E., Molina-Arcas M., Krygowska A. A., East P., Warne P., Nicol A. and Downward J. (2016). RAS signalling through PI3-Kinase controls cell migration via modulation of Reelin expression. Nat. Commun. 7, 11245 10.1038/ncomms11245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantaravisoot N., Wongkongkathep P., Loo J. A., Mischel P. S. and Tamanoi F. (2015). Significance of filamin A in mTORC2 function in glioblastoma. Mol. Cancer 14, 127 10.1186/s12943-015-0396-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charest P. G. G. and Firtel R. A. A. (2007). Big roles for small GTPases in the control of directed cell movement. Biochem. J. 401, 377-390. 10.1042/BJ20061432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charest P. G. G., Shen Z., Lakoduk A., Sasaki A. T. T., Briggs S. P. P. and Firtel R. A. A. (2010). A Ras signaling complex controls the RasC-TORC2 pathway and directed cell migration. Dev. Cell 18, 737-749. 10.1016/j.devcel.2010.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Xu B., Liu L., Liu C., Luo Y., Chen X., Barzegar M., Chung J. and Huang S. (2015a). Both mTORC1 and mTORC2 are involved in the regulation of cell adhesion. Oncotarget 6, 7136-7150. 10.18632/oncotarget.3044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Cheng H., Pan T., Liu Y., Su Y., Ren C., Huang D., Zha X. and Liang C. (2015b). mTOR regulate EMT through RhoA and Rac1 pathway in prostate cancer. Mol. Carcinog. 54, 1086-1095. 10.1002/mc.22177 [DOI] [PubMed] [Google Scholar]
- Chen X., Liu M., Tian Y., Li J., Qi Y., Zhao D., Wu Z., Huang M., Wong C. C. L., Wang H.-W. et al. (2018). Cryo-EM structure of human mTOR complex 2. Cell Res. 28, 518-528. 10.1038/s41422-018-0029-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comer F. I. and Parent C. A. (2006). Phosphoinositide 3-kinase activity controls the chemoattractant-mediated activation and adaptation of adenylyl cyclase. Mol. Biol. Cell 17, 357-366. 10.1091/mbc.e05-08-0781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comer F. I., Lippincott C. K., Masbad J. J. and Parent C. A. (2005). The PI3K-mediated activation of CRAC independently regulates adenylyl cyclase activation and chemotaxis. Curr. Biol. 15, 134-139. 10.1016/j.cub.2005.01.007 [DOI] [PubMed] [Google Scholar]
- Ebner M., Sinkovics B., Szczygieł M., Ribeiro D. W. and Yudushkin I. (2017). Localization of mTORC2 activity inside cells. J. Cell Biol. 216, 343-353. 10.1083/jcb.201610060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eller-Borges R., Batista W. L., da Costa P. E., Tokikawa R., Curcio M. F., Strumillo S. T., Sartori A., Moraes M. S., de Oliveira G. A., Taha M. O. et al. (2015). Ras, Rac1, and phosphatidylinositol-3-kinase (PI3K) signaling in nitric oxide induced endothelial cell migration. Nitric Oxide 47, 40-51. 10.1016/j.niox.2015.03.004 [DOI] [PubMed] [Google Scholar]
- Engin H. B., Carlin D., Pratt D. and Carter H. (2017). Modeling of RAS complexes supports roles in cancer for less studied partners. BMC Biophysics 10, 5 10.1186/s13628-017-0037-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facchinetti V., Ouyang W., Wei H., Soto N., Lazorchak A., Gould C., Lowry C., Newton A. C., Mao Y., Miao R. Q. et al. (2008). The mammalian target of rapamycin complex 2 controls folding and stability of Akt and protein kinase C. EMBO J. 27, 1932-1943. 10.1038/emboj.2008.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhan M. A., Carmine-Simmen K., Lewis J. D., Moore R. B. and Murray A. G. (2015). Endothelial cell mTOR complex-2 regulates sprouting angiogenesis. PLoS ONE 10, e0135245 10.1371/journal.pone.0135245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster D. A. (2013). Phosphatidic acid and lipid-sensing by mTOR. Trends Endocrinol. Metab. 24, 272-278. 10.1016/j.tem.2013.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frias M. A., Thoreen C. C., Jaffe J. D., Schroder W., Sculley T., Carr S. A. and Sabatini D. M. (2006). mSin1 is necessary for Akt/PKB phosphorylation, and its isoforms define three distinct mTORC2s. Curr. Biol. 16, 1865-1870. 10.1016/j.cub.2006.08.001 [DOI] [PubMed] [Google Scholar]
- Fruman D. A., Chiu H., Hopkins B. D., Bagrodia S., Cantley L. C. and Abraham R. T. (2017). The PI3K pathway in human disease. Cell 170, 605-635. 10.1016/j.cell.2017.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funamoto S., Milan K., Meili R. and Firtel R. A. (2001). Role of phosphatidylinositol 3′ kinase and a downstream pleckstrin homology domain-containing protein in controlling chemotaxis in Dictyostelium. J. Cell Biol. 153, 795-810. 10.1083/jcb.153.4.795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funamoto S., Meili R., Lee S., Parry L. and Firtel R. A. (2002). Spatial and temporal regulation of 3-phosphoinositides by PI 3-kinase and PTEN mediates chemotaxis. Cell 109, 611-623. 10.1016/S0092-8674(02)00755-9 [DOI] [PubMed] [Google Scholar]
- Gan X., Wang J., Su B. and Wu D. (2011). Evidence for direct activation of mTORC2 kinase activity by phosphatidylinositol 3,4,5-trisphosphate. J. Biol. Chem. 286, 10998-11002. 10.1074/jbc.M110.195016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkountakos A., Pilotto S., Mafficini A., Vicentini C., Simbolo M., Milella M., Tortora G., Scarpa A., Bria E. and Corbo V. (2018). Unmasking the impact of Rictor in cancer: novel insights of mTORC2 complex. Carcinogenesis 39, 971-980. 10.1093/carcin/bgy086 [DOI] [PubMed] [Google Scholar]
- Gleason C. E., Oses-Prieto J. A., Li K. H., Saha B., Situ G., Burlingame A. L. and Pearce D. (2019). Phosphorylation at distinct subcellular locations underlies specificity in mTORC2 activation of SGK1 and Akt. J. Cell Sci. 132, jcs.224931 10.1242/jcs.224931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruver J. S., Wikswo J. P. Jr and Chung C. Y. (2008). 3′-phosphoinositides regulate the coordination of speed and accuracy during chemotaxis. Biophys. J. 95, 4057-4067. 10.1529/biophysj.108.130179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulhati P., Bowen K. A., Liu J., Stevens P. D., Rychahou P. G., Chen M., Lee E. Y., Weiss H. L., O'Connor K. L., Gao T. et al. (2011). mTORC1 and mTORC2 regulate EMT, motility, and metastasis of colorectal cancer via RhoA and Rac1 signaling pathways. Cancer Res. 71, 3246-3256. 10.1158/0008-5472.CAN-10-4058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Li D., Cook S. L., Yoon M.-S., Kapoor A., Rao C. V., Kenis P. J. A., Chen J. and Wang F. (2013). Mammalian target of rapamycin and Rictor control neutrophil chemotaxis by regulating Rac/Cdc42 activity and the actin cytoskeleton. Mol. Biol. Cell 24, 3369-3380. 10.1091/mbc.e13-07-0405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer F., Fields S., Schneider C. and Martin G. S. (1994). Activated Ras interacts with the Ral guanine nucleotide dissociation stimulator. Proc. Natl. Acad. Sci. USA 91, 11089-11093. 10.1073/pnas.91.23.11089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holroyd A. K. and Michie A. M. (2018). The role of mTOR-mediated signaling in the regulation of cellular migration. Immunol. Lett. 196, 74-79. 10.1016/j.imlet.2018.01.015 [DOI] [PubMed] [Google Scholar]
- Hoxhaj G. and Manning B. D. (2019). The PI3K–AKT network at the interface of oncogenic signalling and cancer metabolism. Nat. Rev. Cancer 20, 74-88. 10.1038/s41568-019-0216-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikenoue T., Inoki K., Yang Q., Zhou X. and Guan K.-L. (2008). Essential function of TORC2 in PKC and Akt turn motif phosphorylation, maturation and signalling. EMBO J. 27, 1919-1931. 10.1038/emboj.2008.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto E., Loewith R., Schmidt A., Lin S., Rüegg M. A., Hall A. and Hall M. N. (2004). Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat. Cell Biol. 6, 1122-1128. 10.1038/ncb1183 [DOI] [PubMed] [Google Scholar]
- Jacinto E., Facchinetti V., Liu D., Soto N., Wei S., Jung S. Y., Huang Q., Qin J. and Su B. (2006). SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell 127, 125-137. 10.1016/j.cell.2006.08.033 [DOI] [PubMed] [Google Scholar]
- Jaiswal P., Majithia A. R., Rosel D., Liao X.-H., Khurana T. and Kimmel A. R. (2019). Integrated actions of mTOR complexes 1 and 2 for growth and development of Dictyostelium. Int. J. Dev. Biol. 63, 521-527. 10.1387/ijdb.190245ak [DOI] [PubMed] [Google Scholar]
- Janetopoulos C., Borleis J., Vazquez F., Iijima M. and Devreotes P. (2005). Temporal and spatial regulation of phosphoinositide signaling mediates cytokinesis. Dev. Cell 8, 467-477. 10.1016/j.devcel.2005.02.010 [DOI] [PubMed] [Google Scholar]
- Jean S. and Kiger A. A. (2014). Classes of phosphoinositide 3-kinases at a glance. J. Cell Sci. 127, 923-928. 10.1242/jcs.093773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhanwar-Uniyal M., Wainwright J. V., Mohan A. L., Tobias M. E., Murali R., Gandhi C. D. and Schmidt M. H. (2019). Diverse signaling mechanisms of mTOR complexes: mTORC1 and mTORC2 in forming a formidable relationship. Adv. Biol. Regul. 72, 51-62. 10.1016/j.jbior.2019.03.003 [DOI] [PubMed] [Google Scholar]
- Kamimura Y. and Devreotes P. N. (2010). Phosphoinositide-dependent protein kinase (PDK) activity regulates phosphatidylinositol 3,4,5-trisphosphate-dependent and -independent protein kinase B activation and chemotaxis. J. Biol. Chem. 285, 7938-7946. 10.1074/jbc.M109.089235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimura Y., Xiong Y., Iglesias P. A., Hoeller O., Bolourani P. and Devreotes P. N. (2008). PIP3-independent activation of TorC2 and PKB at the cell's leading edge mediates chemotaxis. Curr. Biol. 18, 1034-1043. 10.1016/j.cub.2008.06.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karuppasamy M., Kusmider B., Oliveira T. M., Gaubitz C., Prouteau M., Loewith R. and Schaffitzel C. (2017). Cryo-EM structure of Saccharomyces cerevisiae target of rapamycin complex 2. Nat. Commun. 8, 1729 10.1038/s41467-017-01862-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan I., Rhett J. M. and O'Bryan J. P. (2020). Therapeutic targeting of RAS: new hope for drugging the “undruggable”. Biochim. Biophy. Acta– Mol. Cell Res. 1867, 118570 10.1016/j.bbamcr.2019.118570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna A., Lotfi P., Chavan A. J., Montaño N. M., Bolourani P., Weeks G., Shen Z., Briggs S. P., Pots H., Van Haastert P. J. M. et al. (2016). The small GTPases Ras and Rap1 bind to and control TORC2 activity. Sci. Rep. 6, 25823 10.1038/srep25823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim L. C., Cook R. S. and Chen J. (2017). mTORC1 and mTORC2 in cancer and the tumor microenvironment. Oncogene 36, 2191-2201. 10.1038/onc.2016.363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen J. R., Fritzen A. M., James D. E., Jensen T. E., Kleinert M. and Richter E. A. (2020). Growth factor-dependent and -independent activation of mTORC2. Trends Endocrinol. Metab. 31, 13-24. 10.1016/j.tem.2019.09.005 [DOI] [PubMed] [Google Scholar]
- Kodaki T., Woscholski R., Hallberg B., Rodriguez-Viciana P., Downward J. and Parker P. J. (1994). The activation of phosphatidylinositol 3-kinase by Ras. Curr. Biol. 4, 798-806. 10.1016/S0960-9822(00)00177-9 [DOI] [PubMed] [Google Scholar]
- Kölsch V., Charest P. G. and Firtel R. A. (2008). The regulation of cell motility and chemotaxis by phospholipid signaling. J. Cell Sci. 121, 551-559. 10.1242/jcs.023333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalski J. R., Bhaduri A., Zehnder A. M., Neela P. H., Che Y., Wozniak G. G. and Khavari P. A. (2019). The functional proximal proteome of oncogenic ras includes mTORC2. Mol. Cell 73, 830-844.e12. 10.1016/j.molcel.2018.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz J., Schneider U., Howald I., Schmidt A. and Hall M. N. (2000). HEAT repeats mediate plasma membrane localization of Tor2p in yeast. J. Biol. Chem. 275, 37011037020 10.1074/jbc.M007296200 [DOI] [PubMed] [Google Scholar]
- Lee S., Parent C. A., Insall R. and Firtel R. A. (1999). A novel Ras-interacting protein required for chemotaxis and cyclic adenosine monophosphate signal relay in Dictyostelium. Mol. Biol. Cell 10, 2829-2845. 10.1091/mbc.10.9.2829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Comer F. I., Sasaki A., McLeod I. X., Duong Y., Okumura K., Yates J. R. III, Parent C. A. and Firtel R. A. (2005). TOR complex 2 integrates cell movement during chemotaxis and signal relay in Dictyostelium. Mol. Biol. Cell 16, 4572-4583. 10.1091/mbc.e05-04-0342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C. J., Spiegelman G. B. and Weeks G. (2001). RasC is required for optimal activation of adenylyl cyclase and Akt/PKB during aggregation. EMBO J. 20, 4490-4499. 10.1093/emboj/20.16.4490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L. and Parent C. A. (2011). TOR kinase complexes and cell migration. J. Cell Biol. 194, 815-824. 10.1083/jcb.201102090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G. Y. and Sabatini D. M. (2020). mTOR at the nexus of nutrition, growth, ageing and disease. Nat. Rev. Mol. Cell Biol. 21, 183-203. 10.1038/s41580-019-0199-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Das S., Losert W. and Parent C. A. (2010). mTORC2 Regulates neutrophil chemotaxis in a cAMP- and RhoA-dependent fashion. Dev. Cell 19, 845-857. 10.1016/j.devcel.2010.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Gan W., Inuzuka H., Lazorchak A. S., Gao D., Arojo O., Liu D., Wan L., Zhai B., Yu Y. et al. (2013). Sin1 phosphorylation impairs mTORC2 complex integrity and inhibits downstream Akt signaling to suppress tumorigenesis. Nat. Cell Biol. 15, 1340 10.1038/ncb2860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Gan W., Chin Y. R., Ogura K., Guo J., Zhang J., Wang B., Blenis J., Cantley L. C., Toker A. et al. (2015). PtdIns(3,4,5)P3-dependent activation of the mTORC2 kinase complex. Cancer Discov. 5, 1194-1209. 10.1158/2159-8290.CD-15-0460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lone M.-U.-D., Miyan J., Asif M., Malik S. A., Dubey P., Singh V., Singh K., Mitra K., Pandey D., Haq W. et al. (2019). Direct physical interaction of active Ras with mSIN1 regulates mTORC2 signaling. BMC Cancer 19, 1236 10.1186/s12885-019-6422-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long X., Lin Y., Ortiz-Vega S., Yonezawa K. and Avruch J. (2005). Rheb binds and regulates the mTOR kinase. Curr. Biol. 15, 702-713. 10.1016/j.cub.2005.02.053 [DOI] [PubMed] [Google Scholar]
- Mahadeo D. C. and Parent C. A. (2006). Signal relay during the life cycle of dictyostelium. Curr. Top. Dev. Biol., 115-140. 10.1016/S0070-2153(05)73004-0 [DOI] [PubMed] [Google Scholar]
- Malumbres M. and Barbacid M. (2003). RAS oncogenes: the first 30 years. Nat. Rev. Cancer 3, 459-465. 10.1038/nrc1097 [DOI] [PubMed] [Google Scholar]
- Martinez Marshall M. N., Emmerstorfer-Augustin A., Leskoske K. L., Zhang L. H., Li B. and Thorner J. (2019). Analysis of the roles of phosphatidylinositol-4,5- bis phosphate and individual subunits in assembly, localization, and function of Saccharomyces cerevisiae target of rapamycin complex 2. Mol. Biol. Cell 30, 1555-1574. 10.1091/mbc.E18-10-0682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick F. (2019). Progress in targeting RAS with small molecule drugs. Biochem. J. 476, 365-374. 10.1042/BCJ20170441 [DOI] [PubMed] [Google Scholar]
- Menon D., Salloum D., Bernfeld E., Gorodetsky E., Akselrod A., Frias M. A., Sudderth J., Chen P.-H., DeBerardinis R. and Foster D. A. (2017). Lipid sensing by mTOR complexes via de novo synthesis of phosphatidic acid. J. Biol. Chem. 292, 6303-6311. 10.1074/jbc.M116.772988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukaida S., Evans B. A., Bengtsson T., Hutchinson D. S. and Sato M. (2017). Adrenoceptors promote glucose uptake into adipocytes and muscle by an insulin-independent signaling pathway involving mechanistic target of rapamycin complex 2. Pharmacol. Res. 116, 87-92. 10.1016/j.phrs.2016.12.022 [DOI] [PubMed] [Google Scholar]
- Muñoz-Maldonado C., Zimmer Y. and Medová M. (2019). A comparative analysis of individual RAS mutations in cancer biology. Front. Oncol. 9, 1088 10.3389/fonc.2019.01088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers S. A., Han J. W., Lee Y., Firtel R. A. and Chung C. Y. (2005). A Dictyostelium homologue of WASP is required for polarized F-actin assembly during chemotaxis. Mol. Biol. Cell 16, 2191-2206. 10.1091/mbc.e04-09-0844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakhaei-Rad S., Haghighi F., Nouri P., Rezaei Adariani S., Lissy J., Kazemein Jasemi N. S., Dvorsky R. and Ahmadian M. R. (2018). Structural fingerprints, interactions, and signaling networks of RAS family proteins beyond RAS isoforms. Crit. Rev. Biochem. Mol. Biol. 53, 130-156. 10.1080/10409238.2018.1431605 [DOI] [PubMed] [Google Scholar]
- Nussinov R., Tsai C.-J. and Jang H. (2019). Does Ras activate Raf and PI3K allosterically? Front. Oncol. 9, 1231 10.3389/fonc.2019.01231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh W. J. and Jacinto E. (2011). mTOR complex 2 signaling and functions. Cell Cycle 10, 2305-2316. 10.4161/cc.10.14.16586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh W. J., Wu C., Kim S. J., Facchinetti V., Julien L.-A., Finlan M., Roux P. P., Su B. and Jacinto E. (2010). mTORC2 can associate with ribosomes to promote cotranslational phosphorylation and stability of nascent Akt polypeptide. EMBO J. 29, 3939-3951. 10.1038/emboj.2010.271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrem J. M., Peters U., Sos M. L., Wells J. A. and Shokat K. M. (2013). K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 503, 548-551. 10.1038/nature12796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partovian C., Ju R., Zhuang Z. W., Martin K. A. and Simons M. (2008). Syndecan-4 regulates subcellular localization of mTOR complex2 and Akt activation in a PKCα-dependent manner in endothelial cells. Mol. Cell 32, 140-149. 10.1016/j.molcel.2008.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson T. R., Laplante M., Thoreen C. C., Sancak Y., Kang S. A., Kuehl W. M., Gray N. S. and Sabatini D. M. (2009). DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell 137, 873-886. 10.1016/j.cell.2009.03.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyrollier K., Hajdych E., Gray A., Litherland G. J., Prescott A. R., Leslie N. R. and Hundal H. S. (2000). A role for the actin cytoskeleton in the hormonal and growth-factor-mediated activation of protein kinase B. Biochem. J. 352, 617-622. 10.1042/bj3520617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma M., Roelofs J., Goedhart J., Loovers H. M., Visser A. J. W. G. and Van Haastert P. J. M. (2004). Sensitization of Dictyostelium chemotaxis by phosphoinositide-3-kinase-mediated self-organizing signalling patches. J. Cell Sci. 117, 2925-2935. 10.1242/jcs.01143 [DOI] [PubMed] [Google Scholar]
- Pylayeva-Gupta Y., Grabocka E. and Bar-Sagi D. (2011). RAS oncogenes: weaving a tumorigenic web. Nat. Rev. Cancer 11, 761-774. 10.1038/nrc3106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinaswamy M. K. and Burke J. E. (2020). Class I phosphoinositide 3-kinase (PI3K) regulatory subunits and their roles in signaling and disease. Adv. Biol. Regul. 75, 100657 10.1016/j.jbior.2019.100657 [DOI] [PubMed] [Google Scholar]
- Riehle R. D., Cornea S. and Degterev A. (2013). Role of phosphatidylinositol 3,4,5-trisphosphate in cell signaling. Adv. Exp. Med. Biol. 991, 105-139. 10.1007/978-94-007-6331-9_7 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Viciana P., Warne P. H., Dhand R., Vanhaesebroeck B., Gout I., Fry M. J., Waterfield M. D. and Downward J. (1994). Phosphatidylinositol-3-OH kinase as a direct target of Ras. Nature 370, 527-532. 10.1038/370527a0 [DOI] [PubMed] [Google Scholar]
- Roelants F. M., Leskoske K. L., Marshall M. N. M., Locke M. N. and Thorner J. (2017). The TORC2-dependent signaling network in the yeast Saccharomyces cerevisiae. Biomolecules 7, 66 10.3390/biom7030066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosel D., Khurana T., Majithia A., Huang X., Bhandari R. and Kimmel A. R. (2012). TOR complex 2 (TORC2) in Dictyostelium suppresses phagocytic nutrient capture independently of TORC1-mediated nutrient sensing. J. Cell Sci. 125, 37-48. 10.1242/jcs.077040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saci A., Cantley L. C. and Carpenter C. L. (2011). Rac1 regulates the activity of mTORC1 and mTORC2 and controls cellular size. Mol. Cell 42, 50-61. 10.1016/j.molcel.2011.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov D. D., Guertin D. A., Ali S. M. and Sabatini D. M. (2005). Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307, 1098-1101. 10.1126/science.1106148 [DOI] [PubMed] [Google Scholar]
- Sasaki A. T., Chun C., Takeda K. and Firtel R. A. (2004). Localized Ras signaling at the leading edge regulates PI3K, cell polarity, and directional cell movement. J. Cell Biol. 167, 505-518. 10.1083/jcb.200406177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A. T., Janetopoulos C., Lee S., Charest P. G., Takeda K., Sundheimer L. W., Meili R., Devreotes P. N. and Firtel R. A. (2007). G protein-independent Ras/PI3K/F-actin circuit regulates basic cell motility. J. Cell Biol. 178, 185-191. 10.1083/jcb.200611138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M., Dehvari N., Oberg A. I., Dallner O. S., Sandström A. L., Olsen J. M., Csikasz R. I., Summers R. J., Hutchinson D. S. and Bengtsson T. (2014). Improving type 2 diabetes through a distinct adrenergic signaling pathway involving mTORC2 that mediates glucose uptake in skeletal muscle. Diabetes 63, 4115-4129. 10.2337/db13-1860 [DOI] [PubMed] [Google Scholar]
- Sato T., Ishii J., Ota Y., Sasaki E., Shibagaki Y. and Hattori S. (2016). Mammalian target of rapamycin (mTOR) complex 2 regulates filamin A-dependent focal adhesion dynamics and cell migration. Genes Cells 21, 579-593. 10.1111/gtc.12366 [DOI] [PubMed] [Google Scholar]
- Schroder W. A., Buck M., Cloonan N., Hancock J. F., Suhrbier A., Sculley T. and Bushell G. (2007). Human Sin1 contains Ras-binding and pleckstrin homology domains and suppresses Ras signalling. Cell. Signal. 19, 1279-1289. 10.1016/j.cellsig.2007.01.013 [DOI] [PubMed] [Google Scholar]
- Senoo H., Kamimura Y., Kimura R., Nakajima A., Sawai S., Sesaki H. and Iijima M. (2019). Phosphorylated Rho–GDP directly activates mTORC2 kinase towards AKT through dimerization with Ras–GTP to regulate cell migration. Nat. Cell Biol. 21, 867-878. 10.1038/s41556-019-0348-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simanshu D. K., Nissley D. V. and McCormick F. (2017). RAS Proteins and their regulators in human disease. Cell 170, 17-33. 10.1016/j.cell.2017.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smrz D., Cruse G., Beaven M. A., Kirshenbaum A., Metcalfe D. D. and Gilfillan A. M. (2014). Rictor negatively regulates high-affinity receptors for IgE-induced mast cell degranulation. J. Immunol. 193, 5924-5932. 10.4049/jimmunol.1303495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokoe D., Stephens L. R., Copeland T., Gaffney P. R., Reese C. B., Painter G. F., Holmes A. B., McCormick F. and Hawkins P. T. (1997). Dual role of phosphatidylinositol-3,4,5-trisphosphate in the activation of protein kinase B. Science (New York, N.Y.) 277, 567-570. 10.1126/science.277.5325.567 [DOI] [PubMed] [Google Scholar]
- Strahl T. and Thorner J. (2007). Synthesis and function of membrane phosphoinositides in budding yeast, Saccharomyces cerevisiae. Biochim. Biophys. Acta. 1771, 353-404. 10.1016/j.bbalip.2007.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill T. W., Cohen A., Diefenbacher M., Trautwein M., Martin D. E. and Hall M. N. (2008). TOR1 and TOR2 have distinct locations in live cells. Eukaryot. Cell 7, 1819-1830. 10.1128/EC.00088-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuttfeld E., Aylett C. H., Imseng S., Boehringer D., Scaiola A., Sauer E., Hall M. N., Maier T. and Ban N. (2018). Architecture of the human mTORC2 core complex. eLife 7, e33101 10.7554/eLife.33101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suire S., Condliffe A. M., Ferguson G. J., Ellson C. D., Guillou H., Davidson K., Welch H., Coadwell J., Turner M., Chilvers E. R. et al. (2006). Gβγs and the Ras binding domain of p110γ are both important regulators of PI(3)Kγ signalling in neutrophils. Nat. Cell Biol. 8, 1303-1309. 10.1038/ncb1494 [DOI] [PubMed] [Google Scholar]
- Takeda K., Sasaki A. T., Ha H., Seung H. A. and Firtel R. A. (2007). Role of PI3 kinases in chemotaxis in Dictyostelium. J. Biol. Chem. 282, 11874-11884. 10.1074/jbc.M610984200 [DOI] [PubMed] [Google Scholar]
- Tanski W., Roztocil E. and Davies M. G. (2002). Sphingosine-1-phosphate induces Gαi-coupled, PI3K/ras-dependent smooth muscle cell migration. J. Surg. Res. 108, 98-106. 10.1006/jsre.2002.6529 [DOI] [PubMed] [Google Scholar]
- Tatebe H., Murayama S., Yonekura T., Hatano T., Richter D., Furuya T., Kataoka S., Furuita K., Kojima C. and Shiozaki K. (2017). Substrate specificity of tor complex 2 is determined by a ubiquitin-fold domain of the sin1 subunit. eLife 6, e19594 10.7554/eLife.19594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevathasan J. V., Tan E., Hui Z., Lin Y. C., Li Y., Inoue T. and Fivaz M. (2013). The small GTPase HRas shapes local PI3K signals through positive feedback and regulates persistent membrane extension in migrating fibroblasts. Mol. Biol. Cell 24, 2228-2237. 10.1091/mbc.e12-12-0905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton C., Yaka R., Dinh S. and Ron D. (2003). H-Ras modulates N-methyl-D-aspartate receptor function via inhibition of Src tyrosine kinase activity. J. Biol. Chem. 278, 23823-23829. 10.1074/jbc.M302389200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhaesebroeck B. and Alessi D. R. (2000). The PI3K-PDK1 connection: more than just a road to PKB. Biochem. J. 346, 561-576. 10.1042/bj3460561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhaesebroeck B., Guillermet-Guibert J., Graupera M. and Bilanges B. (2010). The emerging mechanisms of isoform-specific PI3K signalling. Nat. Rev. Mol. Cell Biol. 11, 329-341. 10.1038/nrm2882 [DOI] [PubMed] [Google Scholar]
- Vigil D., Cherfils J., Rossman K. L. and Der C. J. (2010). Ras superfamily GEFs and GAPs: validated and tractable targets for cancer therapy? Nat. Rev. Cancer 10, 842-857. 10.1038/nrc2960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojtek A. B., Hollenberg S. M. and Cooper J. A. (1993). Mammalian Ras interacts directly with the serine/threonine kinase raf. Cell 74, 205-214. 10.1016/0092-8674(93)90307-C [DOI] [PubMed] [Google Scholar]
- Wang F., Herzmark P., Weiner O. D., Srinivasan S., Servant G. and Bourne H. R. (2002). Lipid products of PI(3)Ks maintain persistent cell polarity and directed motility in neutrophils. Nat. Cell Biol. 4, 513-518. 10.1038/ncb810 [DOI] [PubMed] [Google Scholar]
- Wedaman K. P., Reinke A., Anderson S., Yates J., Michael McCaffery J. and Powers T. (2003). Tor kinases are in distinct membrane-associated protein complexes in Saccharomyces cerevisiae. Mol. Biol. Cell 14, 1204-1220. 10.1091/mbc.e02-09-0609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlgemuth S., Kiel C., Krämer A., Serrano L., Wittinghofer F., Herrmann C. and Kramer A. (2005). Recognizing and defining true ras binding domains i: biochemical analysis. J. Mol. Biol. 348, 741-758. 10.1016/j.jmb.2005.02.048 [DOI] [PubMed] [Google Scholar]
- Xie J., Wang X. and Proud C. G. (2018). Who does TORC2 talk to? Biochem. J. 475, 1721-1738. 10.1042/BCJ20180130 [DOI] [PubMed] [Google Scholar]
- Yang Q., Inoki K., Ikenoue T. and Guan K. L. (2006). Identification of Sin1 as an essential TORC2 component required for complex formation and kinase activity. Genes Dev. 20, 2820-2832. 10.1101/gad.1461206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H. W., Shin M.-G., Lee S., Kim J.-R., Park W. S., Cho K.-H., Meyer T. and Heo W. D. (2012). Cooperative activation of PI3K by Ras and Rho family small GTPases. Mol. Cell 47, 281-290. 10.1016/j.molcel.2012.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G., Murashige D. S. S., Humphrey S. J. J. and James D. E. E. (2015). A Positive Feedback Loop between Akt and mTORC2 via SIN1 Phosphorylation. Cell Reports 12, 937-943. 10.1016/j.celrep.2015.07.016 [DOI] [PubMed] [Google Scholar]
- Yang H., Jiang X., Li B., Yang H. J., Miller M., Yang A., Dhar A. and Pavletich N. P. N. P. (2017). Mechanisms of mTORC1 activation by RHEB and inhibition of PRAS40. Nature 552, 368-373. 10.1038/nature25023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C.-A., Ortiz-Vega S., Sun Y.-Y., Chien C.-T., Chuang J.-H. and Lin Y. (2017). Association of mSin1 with mTORC2 Ras and Akt reveals a crucial domain on mSin1 involved in Akt activation. Oncotarget 8, 63392-63404. 10.18632/oncotarget.18818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Hua H., Li M., Liu S., Kong Q., Shao T., Wang J., Luo Y., Wang Q., Luo T. et al. (2016). mTORC2 promotes type I insulin-like growth factor receptor and insulin receptor activation through the tyrosine kinase activity of mTOR. Cell Res. 26, 46-65. 10.1038/cr.2015.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Charest P. G. G. and Firtel R. A. A. (2008). Spatiotemporal regulation of Ras activity provides directional sensing. Curr. Biol. 18, 1587-1593. 10.1016/j.cub.2008.08.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H. and Huang S. (2010). Role of mTOR signaling in tumor cell motility, invasion and metastasis. Curr. Protein Pept Sci. 12, 30-42. 10.2174/138920311795659407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Prakash P., Gorfe A. A. and Hancock J. F. (2018). Ras and the plasma membrane: a complicated relationship. Cold Spring Harb. Perspect. Med. 8, a031831 10.1101/cshperspect.a031831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinzalla V., Stracka D., Oppliger W. and Hall M. N. (2011). Activation of mTORC2 by association with the ribosome. Cell 144, 757-768. 10.1016/j.cell.2011.02.014 [DOI] [PubMed] [Google Scholar]
- Zou Z., Chen J., Yang J. and Bai X. (2016). Targeted inhibition of Rictor/mTORC2 in cancer treatment: a new era after Rapamycin. Curr. Cancer Drug Targets 16, 288-304. 10.2174/1568009616666151113120830 [DOI] [PubMed] [Google Scholar]