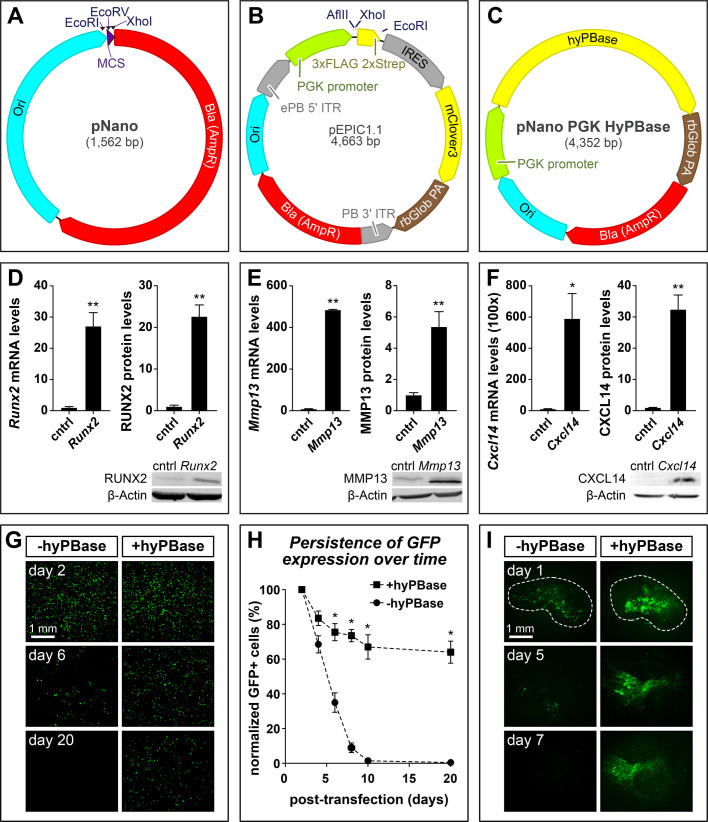
Fig. 1.
Plasmid maps and over-expression analyses. (A) Map of the pNano minimal cloning vector showing restriction sites for cloning, multicloning sites (MCS) in purple, bacterial origin of replication (Ori) in cyan, and bacterial β-lactamase (Bla) resistance gene (AmpR) in red. (B) Map of the pEPIC1.1 piggyBac-integrating constitutively-active expression vector showing piggyBac ITRs and IRES sequences in grey, PGK promoter sequences in green, terminator sequences in brown, and coding sequences in yellow. The pEPIC1.1 vector constitutively expresses mClover3, a GFP. (C) Map of the pNano-hyPBase expression vector used to integrate piggyBac sequences into host genome. (D) Over-expression of Runx2, (E) Mmp13, and (F) Cxcl14 with pEPIC1.1. DF-1 cells were transfected with control (cntrl) empty pEPIC1.1 or pEPIC1.1 plus Runx2, Mmp13, or Cxcl14 coding sequences and harvested 3 days post-transfection. Relative mRNA levels were measured by qPCR and normalized using 18S. Relative protein levels were measured by western blot (WB) and normalized using β-Actin. Representative WBs are shown below. There were four biological replicates for Runx2 and Mmp13, and two for Cxcl14. (G) Fluorescent images showing a time course of DF-1 cells transfected with pEPIC1.1. Cells were transfected either without pNano-hyPBase (left column) or with (right column). Cells were passaged every 2 days and imaged at 2, 6, and 20 days post-transfection. (H) Quantification of GFP positive cells as a fraction of the total number of DF-1 cells transfected with pEPIC1.1 with or without pNano-hyPBase and normalized to 2 days post-transfection. There were two biological replicates for each group. (I) Fluorescent images showing a time course of HH21 chick mandibular primordia electroporated with pEPIC1.1-Cxcl14 either without pNano-hyPBase (left column) or with (right column) cultured, and imaged at day 1, 5, and 7. All qPCR was performed in technical duplicate. A two-tailed t-test was used for all statistical analyses. Error bars represent standard error of the mean (s.e.m.). (*P<0.05; **P<0.005).