Figure 7.
Abnormal Levels of NMJ Markers after Loss of SRSF1
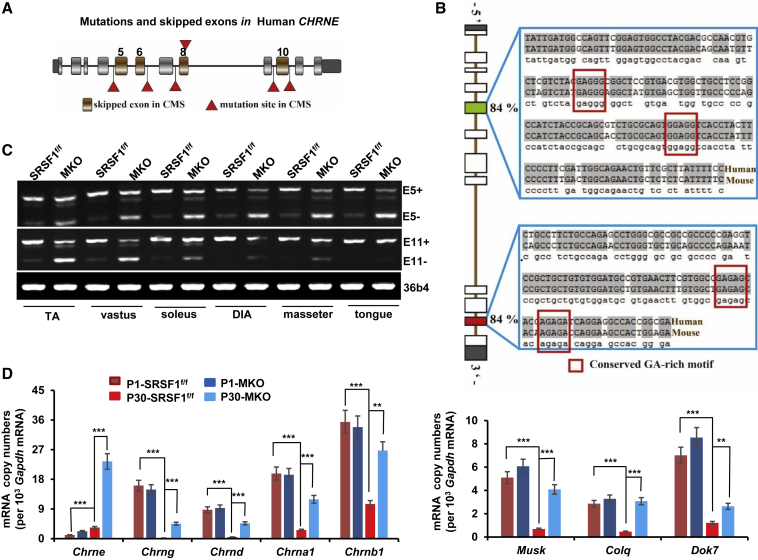
(A) Diagram of human CHRNE gene structure. Brown boxes indicated skipping exons reported in CMS; red triangles indicated point mutations that trigger CMS-related exon skipping. UTRs were marked as black boxes; constitutive exons were marked as gray boxes.
(B) Alignment of human CHRNE E5, E11 and mouse Chrne E5, E11. Note that predicted binding motifs for SRSF1 were conserved between human and mice and are indicated by red rectangles.
(C) Total RNA was isolated from TA, vastus, soleus, DIA, masseter, or tongue muscles from SRSF1f/f and MKO mice at P30. RT-PCR was performed to detect the splicing pattern of Chrne E5 and E11. 36b4 was used as loading control (n = 3).
(D) qPCR was performed using total RNA isolated from hindlimb muscle of P1 mice or from vastus muscles of P30 mice. Comparison of mRNA levels of NMJ markers were performed using mRNA copy numbers (per 103Gapdh mRNA). Error bars depicted mean ± SEM (n = 3).