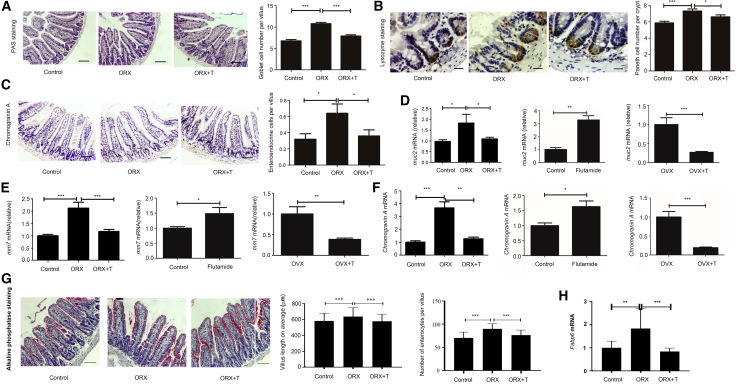
Figure 1.
Androgen's Effect on ISC Lineage Specification
ORX, OVX, and inhibition of AR models were constructed to analyze the change in three secretory lineages and enterocytes. For counting of differentiated cells, 10 villi were randomly chosen for the goblet cells, enteroendocrine cells, and enterocytes, and 10 crypts were randomly captured for the Paneth cells. Ilea were isolated from all mice for immunohistochemistry (IHC), RNA isolation, and quantitative real-time PCR. Data, indicated as mean ± SD, correspond to three independent experiments (A–C, G, n = 6 mice/group/experiment; D–F, H, n = 4 mice/group/experiment).
(A) For the goblet cell count per villus in the ORX model, PAS staining was performed. n = 6. Scale bar, 100 μm.
(B) For the Paneth cell count per crypt in the ORX model, IHC was performed with primary antibody to lysozyme. Scale bar, 50 μm.
(C) For the enteroendocrine cell count per villus in the ORX model, IHC was performed with the primary antibody to chromogranin A. Scale bar, 100 μm.
(D) For determination of relative mRNA levels of muc2 in all groups, quantitative real-time PCR was done.
(E) For determination of relative mRNA levels of mmp7 in all groups, quantitative real-time PCR was done.
(F) For determination of relative mRNA levels of Chromogranin A in all groups, quantitative real-time PCR was done.
(G) For the villus length and enterocyte count per villus in the ORX model, alkaline phosphatase staining was performed. Scale bar, 100 μm.
(H) For determination of relative mRNA levels of Fabp6 in all groups, quantitative real-time PCR was done. ORX+T, supplemented with DHT after ORX.
∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.