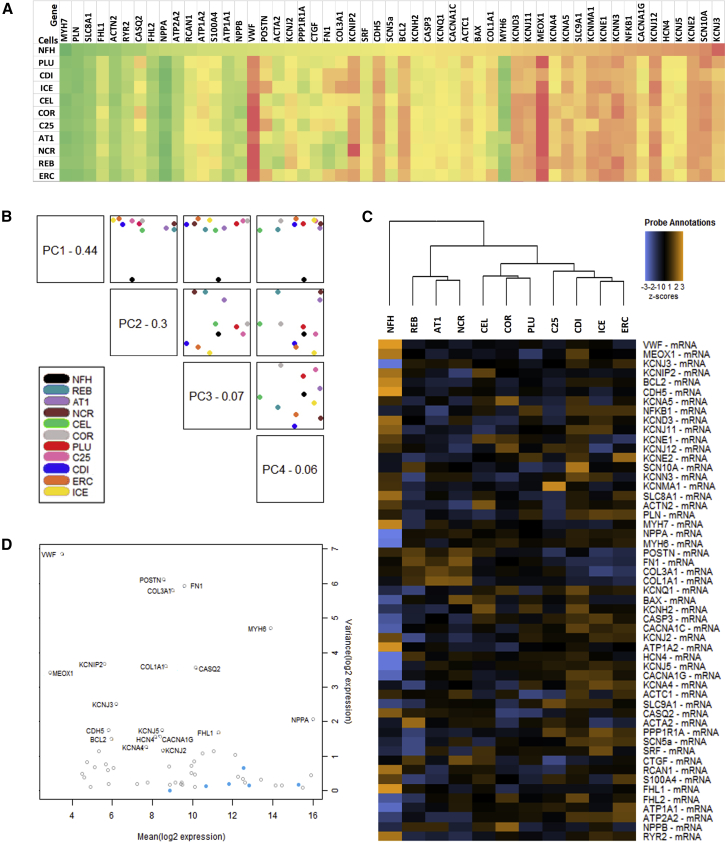
Figure 7.
Gene Expression Analysis of hiPSC-EHTs
Transcriptome analysis (Nanostring) of non-failing human heart (NFH) and the 10 different hiPSC-CM control cell lines in EHT format.
(A) Heatmap of gene expression (log 10) normalized to housekeeping genes and ordered according to NFH levels. Gene expression is color coded with high expression levels marked in green and low gene expression levels in red. See also Figure S5.
(B) Principal-component analysis plotting the first four principal components of the gene expression data against each other.
(C) Hierarchical clustering and dendrogram for the full gene panel. Heatmap of the Z scores of the normalized data, scaled to give all genes equal variance, generated via unsupervised clustering.
(D) Internal quality control plotting variance versus mean normalized signal plot across all lines. Blue dots: housekeeping genes used for normalization. Clear circles: Endogenous genes.