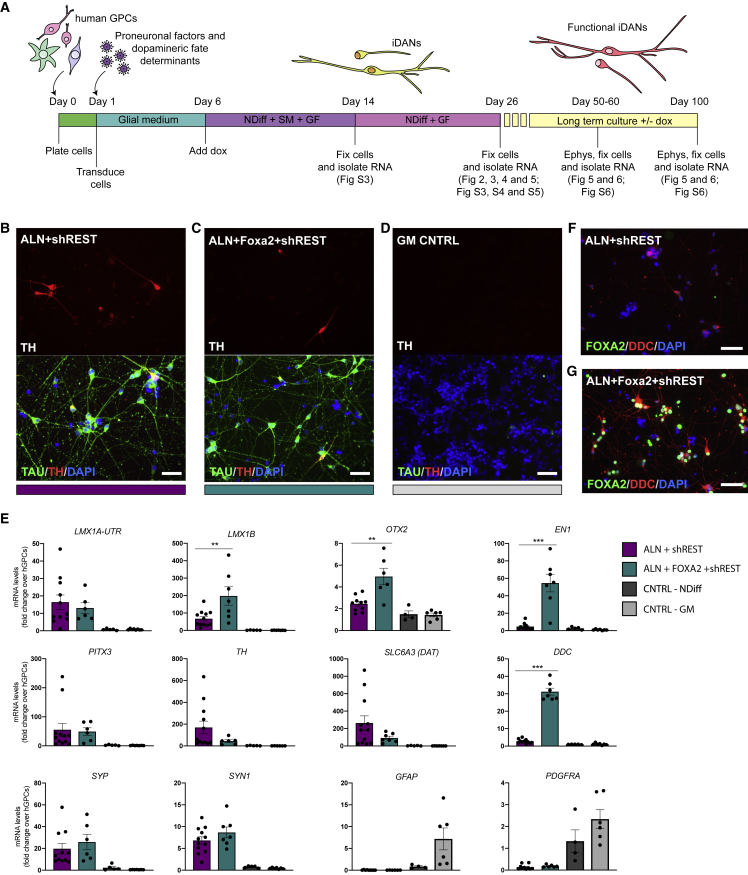
Figure 4.
Direct Conversion of hESC-Derived hGPCs Using Different Combinations of DA Fate Determinants
(A) Schematic overview of the neuronal conversion protocol indicating the different time points of analysis and references to the figures related to each time point.
(B and C) TH+/TAU+ neurons generated with either ALN + shREST (B) or ALN + Foxa2 + shREST (C) 3 weeks after transgene activation starting from hESC-derived hGPCs.
(D) No TH+/TAU+ neurons are generated when the hESC-derived hGPCs are kept for 3 weeks in parallel in glial medium.
(E) qPCR analysis of gene expression of iDANs 3 weeks after transgene activation comparing cells reprogrammed with either ALN + shREST or ALN + Foxa2 + shREST.
(F and G) Immunocytochemical analysis of DDC and FOXA2 in cells reprogrammed using either ALN + shREST (F) or ALN + Foxa2 + shREST (G).
Data are presented as means ± SEM, and all data points have been visualized in the graphs. Each data point represents a replicate from an independent experiment (n = 9–12 for ALN + shREST; n = 6–7 for ALN + Foxa2 + shREST; n = 4–5 for CNTRL-NDIFF; n = 6–8 for CNTRL-GM). The gene expression levels for the conditions ALN + shREST and ALN + Foxa2 + shREST were compared using a Mann-Whitney test; ∗∗p < 0.01 (p = 0.0098 for LMX1B, p = 0.0075 for OTX2); ∗∗∗p < 0.001 (p = 0.0001 for EN1, p < 0.0001 for DDC). Scale bars: 50 μm. See also Figure S5.