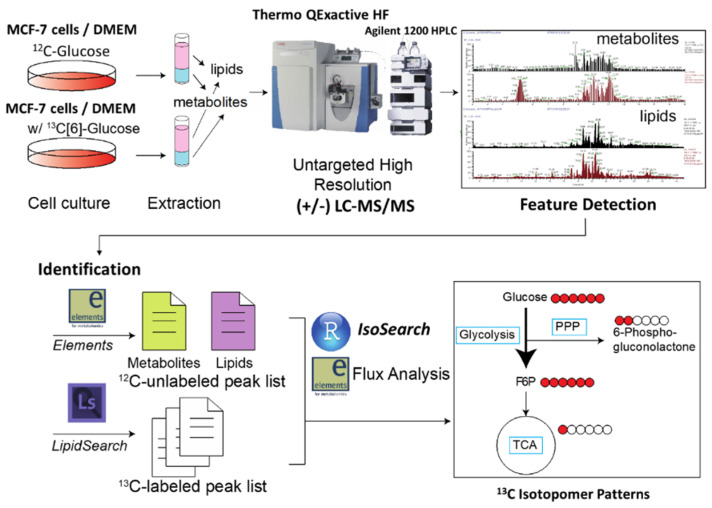
Figure 1.
The workflow of untargeted fluxomic analysis. Two sets of cells are cultured in 12C-glucose or 13C[6]-glucose containing media for isotope labeling. Both non-polar lipids and polar metabolites were extracted using MTBE/methanol, and samples were acquired by high resolution LC-MS/MS via data-dependent acquisition (DDA) with polarity switching. The metabolites and lipids of unlabeled samples were then identified using commercial software and serve as references for labeled samples. The isotopically labeled samples were matched against the references using the in-house R script IsoSearch or the commercial software Scaffold Elements.