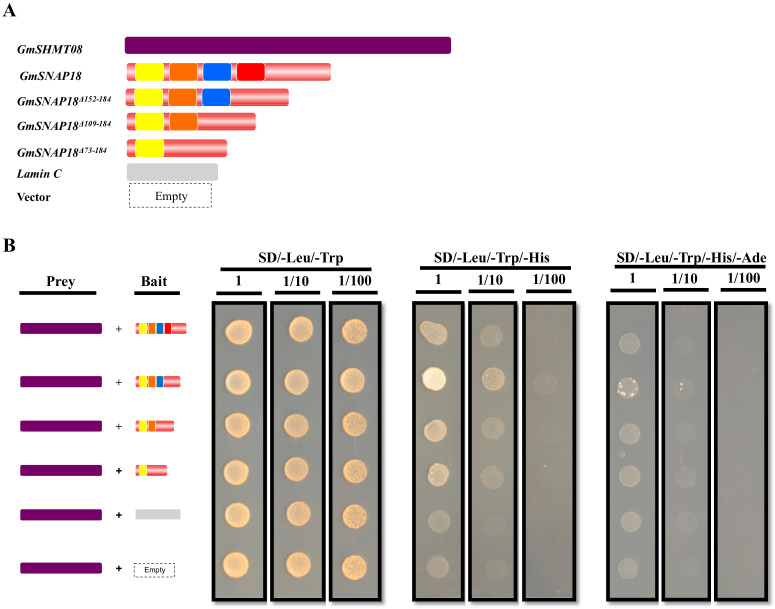
Figure 1.
Impact of mutational analysis of the four TPR motifs GmSNAP18 interaction with GmSHMT08 in yeast. (A) Schematic structures of GmSHMT08 and GmSNAP18 constructs used in the yeast co-transformation assays. (B) Summary of yeast co-transformation assays. Yeast strain AH109 was co-transformed with GmSHMT08 prey vector together with bait constructs containing one (GmSNAP18Δ73-184), two (GmSNAP18Δ109-184), three (GmSNAP18Δ152-184), or four (full length) TPR domains of GmSNAP18. Yeast cells containing bait and prey plasmids were selected by plating the cells on the SD/-Leu/-Trp medium. GmSNAP18/GmSHMT08 interaction were determined by differential growth on the selective SD/Leu/-Trp/-His and SD/-Leu/-Trp/-His/-Ade media. Empty bait plasmid and bait plasmid containing the human Lamin C gene were used as negative controls. Yellow, orange, blue, and red boxes represent TPR1, TPR2, TPR3, and TPR4 Tetratricopeptide repeat motifs at GmSNAP18 predicted protein, respectively. 1, 1/10, and 1/100 represent serial dilutions of the co-transformed yeast cells. The experiment was repeated three times and similar results were obtained.