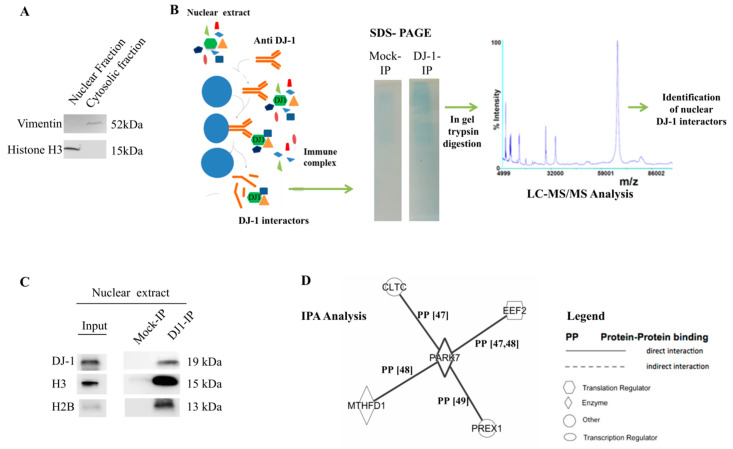
Figure 5.
Mass spectrometry identification of DJ-1 nuclear interactors. (A) The quality of nuclear extracts was assessed by Western blot using vimentin as marker of cytosolic fraction and Histone H3 as marker of nuclear fraction; (B) DJ-1 nuclear interactors were obtained by pull-down nuclear proteins extracted from breast cancer cell (MCF7), using anti-DJ-1 antibody. In parallel, a Mock IP using only protein A/G resin was done to identify unspecific interactors. Immunocomplexes were resolved by SDS-PAGE (Any kD Mini-PROTEAN TGX Precast Protein Gels). Gel bands from mock-IP and DJ-1-IP were manually excised, trypsin digested, and analyzed by LC-MS/MS. Proteins identified in mock-IP were classified as unspecific interactors and eliminated from the list of specific DJ-1 nuclear interactors. The complete list of DJ-1 interactors was provided as Supplementary File S5; (C) The interaction of DJ-1 with Histones H2B and H3 was also confirmed by Western blot analysis. Mock-IP was loaded as negative control; (D) DJ-1 interactors were analyzed using the ingenuity pathways analysis (IPA) tool. Identified networks were provided as Supplementary File S5. IPA analysis allows us to exploit that some of the identified proteins have just been reported as DJ-1 interactors. The network in panel E confirms the direct interaction of DJ-1 with Clathrin heavy chain 1(CLH1), methylenetetrahydrofolate dehydrogenase (MTHFD1), Elongation factor 2 (EF2), and Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein (PREX1). PP on the line connecting proteins indicates protein-protein binding. Peculiar proteins interactions are provided as Supplementary File S5.