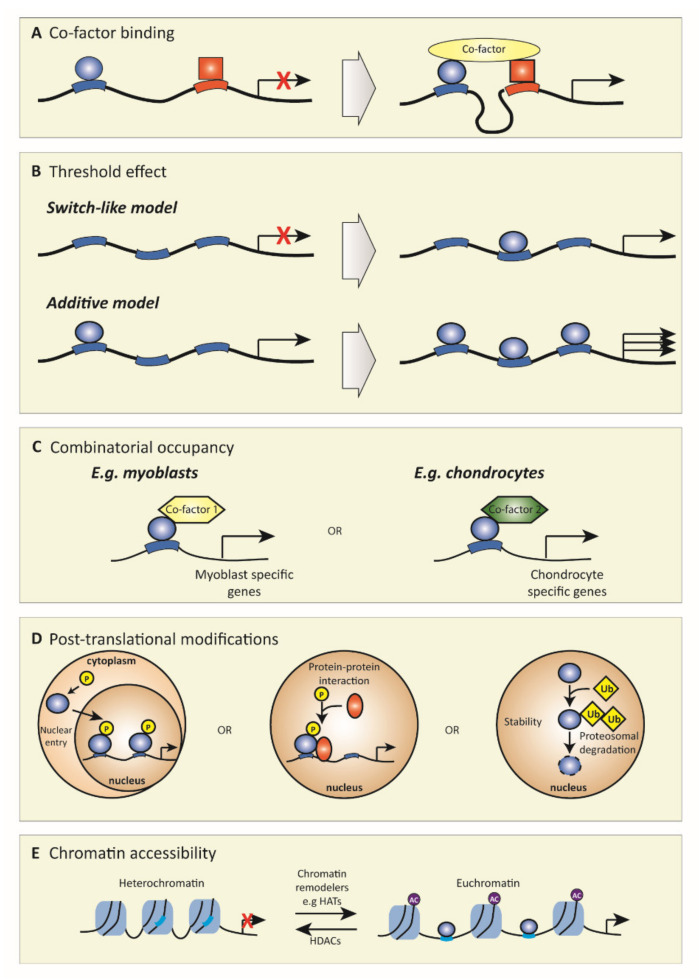
Figure 1.
A schematic depiction of different mechanisms of transcription factor function. Models of how transcription factors can affect transcription by (A) cofactor binding, (B) threshold effect, (C) combinatorial occupancy, (D) post-translation modifications, and (E) chromatin accessibility. (A) Some transcription factors need cofactors to activate transcription. For example, cofactor binding can create a looping of the DNA, which brings transcription factors that are normally far apart into close proximity, which results in activated transcription. (B) For some genes, transcription is an on/off switch where, in the absence of transcription factors, no transcription occurs, while above a certain concentration binding of transcription factors results in transcription (switch-like model). For other genes, binding of more transcription factors results in higher gene expression (additive model). (C) The same transcription factor can induce different responses based on its interaction partners (combinatorial occupancy). For example, in myoblasts, transcription factor 1 interacts with cofactor 1 to express myoblast specific genes, while in chondrocytes, this same transcription factor interacts with cofactor 2 to express chondrocyte specific genes. (D) Post-translational modification is very important for the function of transcription factors, e.g., they can cause nuclear entry of transcription factors that are otherwise rendered in the cytoplasm or they can affect protein–protein interactions, and they have an important role in protein stability and turnover. (E) Chromatin accessibility determines if transcription factor binding sites are available for transcription factors to bind. Heterochromatin, a very dense nucleosome structure, is not accessible for transcription factors to bind, whereas euchromatin, a less dense nucleosome structure, is available for transcription factor binding. HATs = histone acetyl transferases; HDACs = histone deacetylases.