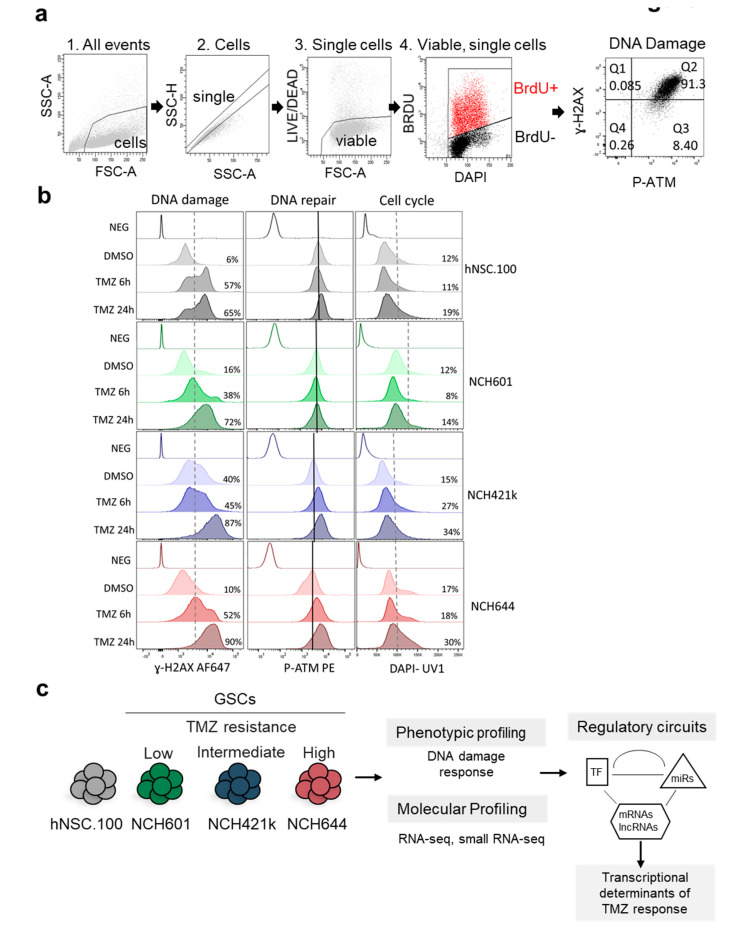
Figure 1.
Glioblastoma (GBM) cells differentially activate DDR (DNA damage Response) in response to TMZ (temozolomide). (a) Gating strategy used for flow cytometry analysis. NCH644 GSCs (Glioblastoma stem-like cells) after 24 h TMZ treatment are shown as an example. (1) Cells were distinguished from debris using Forward Scatter (FSC) and Side Scatter (SSC). (2) Cell doublets and aggregates were gated out based on their properties displayed on the SSC area (SSC-A) versus height (SSC-H) dot plot. (3) Dead cells were recognized by their strong positivity for the dead cell discrimination marker. (4) Proliferating cells were distinguished as BRDU+ (Bromodeoxyuridine / 5-bromo-2’-deoxyuridine) events. DAPI (4′,6-diamidino-2-phenylindole)staining shows cell cycle profile. (5) DNA damage was assessed by measuring the levels of ɣ-H2AX whereas P-ATM (phosphorylated Ataxia telangiectasia mutated) was used to measure the extent of DDR activation. The gating applied discriminates between P-ATM positive vs. negative cells, and ɣ-H2AX high vs. low cells. Percentage of cells in each quartile is presented. (b) Response to TMZ was analyzed in GSCs (NCH601, NCH421k, and NCH644). Neural stem cells (hNSC.100) were used as a non-neoplastic control. DNA damage (ɣ-H2AX), DNA repair (P-ATM), and cell cycle profiles (DAPI) after TMZ treatment (TMZ 6 h and TMZ 24 h) or DMSO as vehicle, are measured in the indicated cell lines. Isotype controls for antibody staining are shown for each cell (Neg). Dotted lines separate low and high ɣ-H2AX cells, or G0/G1 and S/G2/M cell cycle (DAPI graphs). Percentage of ɣ-H2AX high cells and cells in the S/G2/M cell cycle phases is presented. Black lines indicate median expression of P-ATM in DMSO treated cells. (c) GBM stem-like cells of different chemoresistance to TMZ: Low (NCH601), Intermediate (NCH421k), and High (NCH644), and neural stem cells (hNSC.100) were further used for transcriptional profiling and systems level analysis.